Scripting from the history
Overview
Teaching: 0 min
Exercises: 0 minQuestions
How can processing steps be automated from EEGLAB?
How is interactive use of EEGLAB translated to scripts?
How are scripts executed?
Objectives
Understand how to use EEGLAB from the Command Line Interface (CLI) rather than the Graphical User Interface (GUI)
Understand how to automate tasks
Translating interactive EEGLAB procedures into scripts
EEGLAB procedures that are called from the GUI usually save their command line completion to the EEG.history field
Running a procedure on one file in EEGLAB is a good way to explore the data and the process, but once a project goes into production there are several benefits to scripting the procedures. EEGLAB helps with the scripting by storing the command line completion of procedures in the EEG.history field.
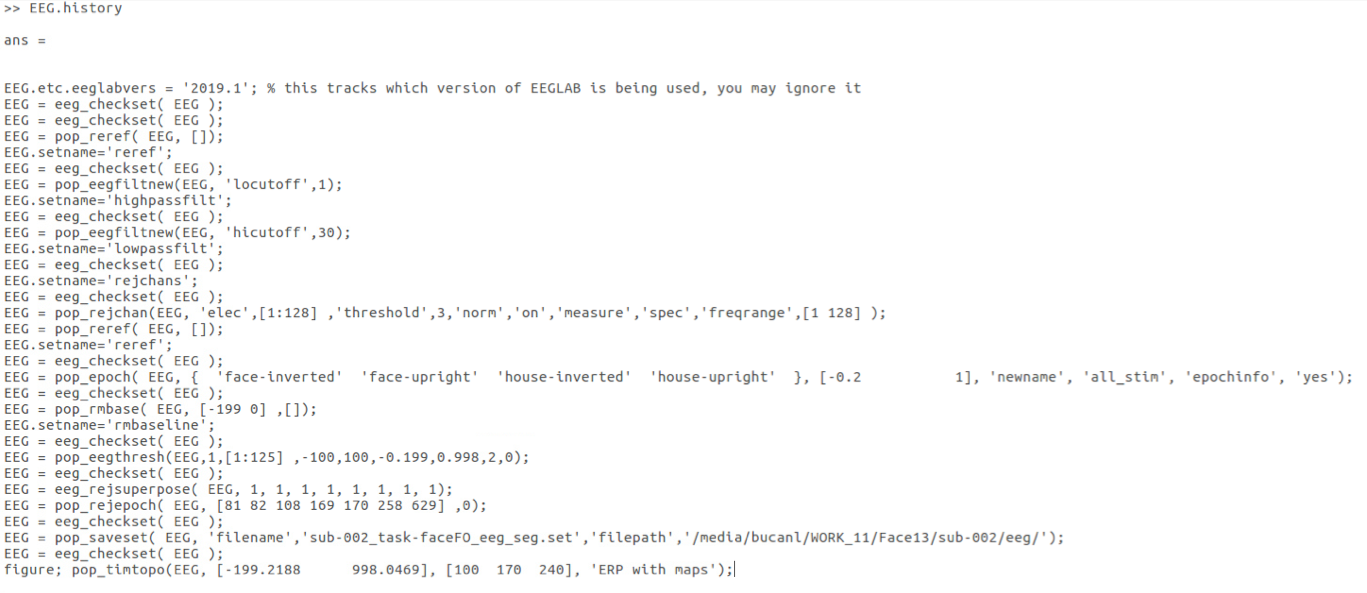
Copy the history string from the Command Window and add it to a script
Use the “edit” function to create a new empty script and open it in the editor:
edit erpproc
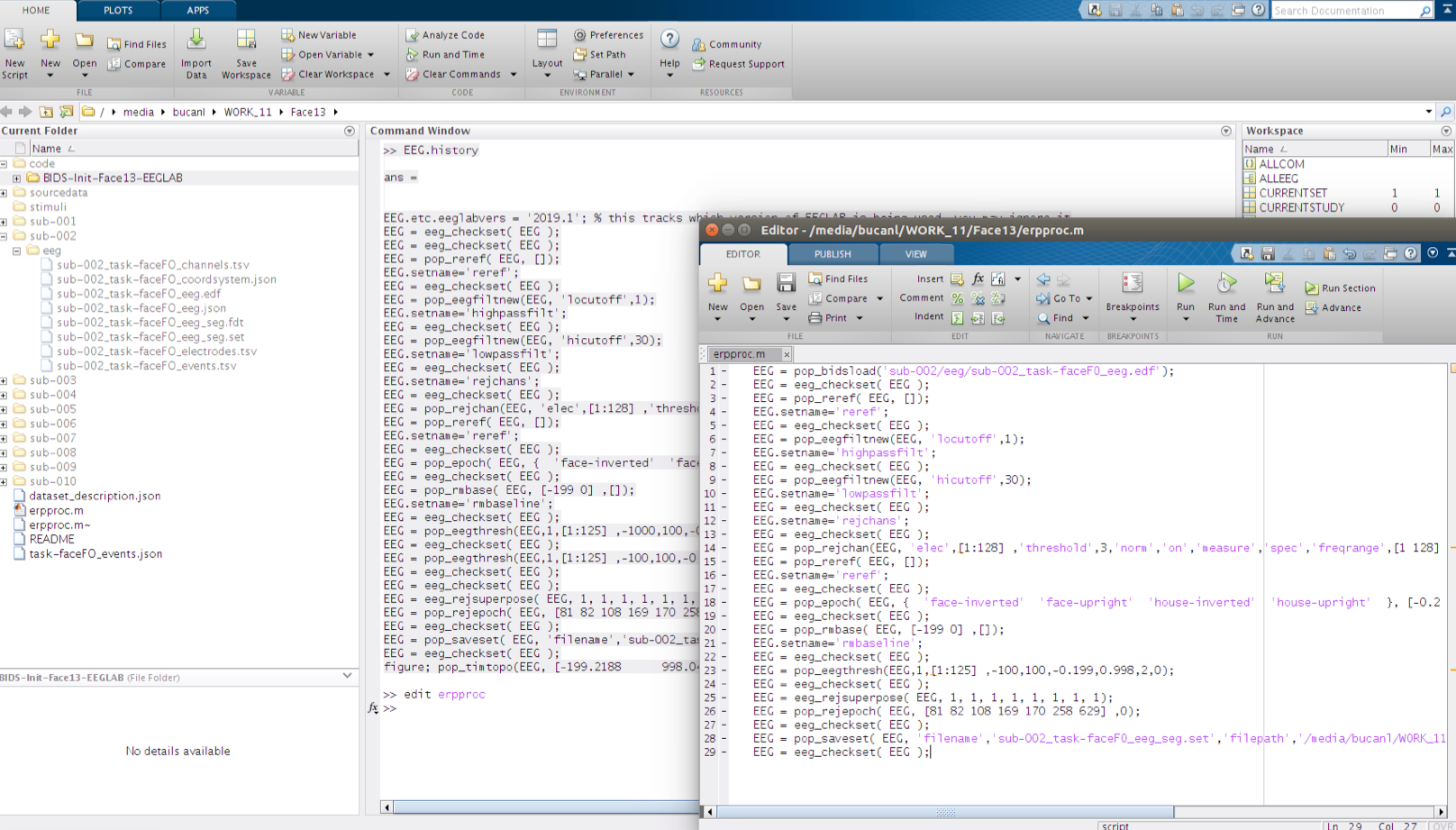
When copying the history from the Command Window there may be several function calls that are not required when running the script. Copy the history string from the first eeg_checkset, up to the point where the processed data was saved to file (pop_saveset). Prior to running the erpproc script, a load command needs to be added at the beginning of the script. To add a load command enter the following code to line 1 of your script:
EEG = pop_bidsload('sub-002/eeg/sub-002_task-faceFO_eeg.edf');
Once that the EEG.history string is pasted to the Editor in “erpproc.m” and the load command has been added, it can be saved then executed from the Command Window by calling its name at the prompt “»”.
>> erpproc
Matlab needs to know where the script is!
Remember that Matlab will execute functions or scripts stored in *.m files if it knows where the file is. This is to say that Matlab needs to be able to find the *.m files if it is going to be able to execute it from the Command Window. By default new scripts are saved to the Current Folder where Matlab sees the scripts by default. If stored elsewhere the directory will need to be included in Matlab’s “path” in order for it to be found.
Editing the script so that it is more generalizable
When EEGLAB stores the Command Line completion of procedures generated from the GUI it does not always store them in a way that is generalizable. Specifically, sometimes there are parameters saved in the function call of the history string that only apply to the specific instance that executed while the history was being created. A good example of this is the pop_rejepoch call in the script.
In fact there are several inputs to functions in this script that may not generalize to other situations (e.g. running a different input file).
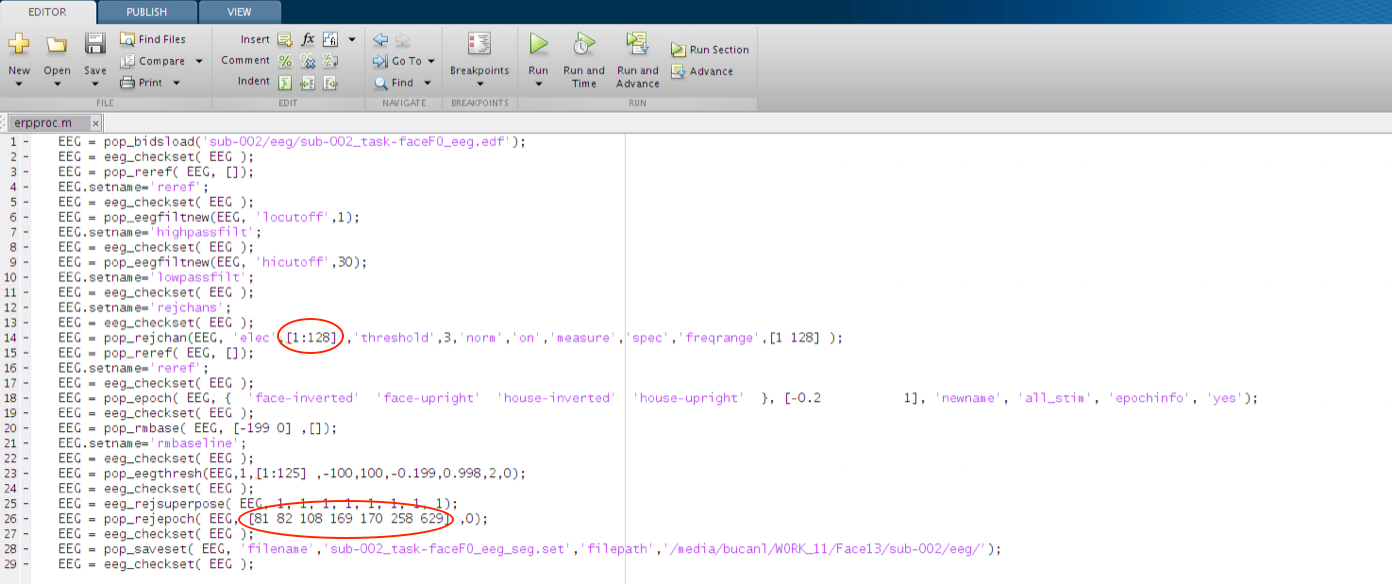
In order to find out what inputs are more appropriate for generalizable calls to the functions we can use the “help” command in the Command Window.
help pop_rejepoch

Once all of the identified input generalizations are identified and alternative parameters are determined the script should look like the following:
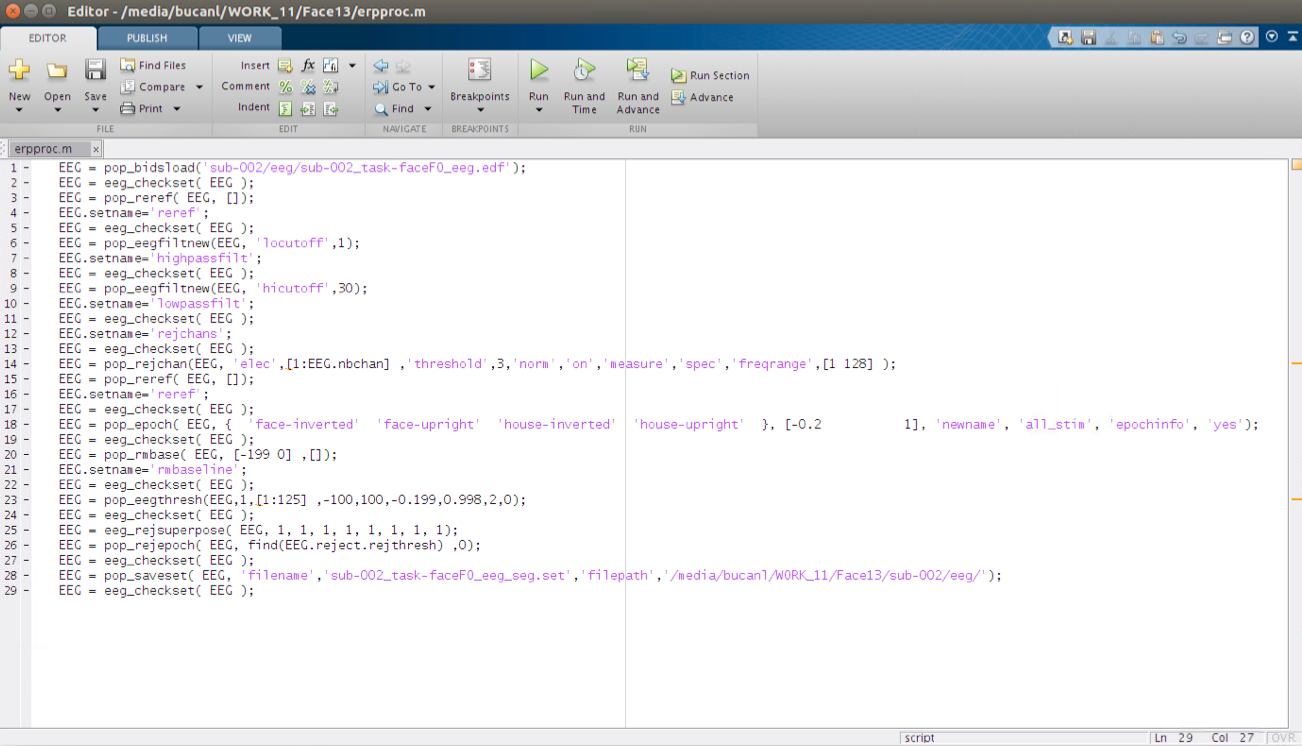
Run the script on multiple files by editing the input and output ‘filename’ parameters
Each participants’ files are saved in a unique location
Given that this example EEG project is stored in accordance with the Brain Imaging Data Structure (BIDS) standard each participant has their files stored within a sub## directory. It is important for this exercise that when editing this script to run different files the ‘filepath’ parameter of the load and save functions are also modified accordingly.
Scripting: How do I love thee, let me count the ways
- Minimize operator error
- Facilitate sharing of procedures
- Provenance, a record of the exact process
- Efficiency
- Process that scales
- etc…
Key Points
Get the computer to work for you in accomplishing EEGLAB procedures