Complex data pipelines
Last updated on 2026-02-03 | Edit this page
Estimated time: 67 minutes
Overview
Questions
- How can I combine everything I’ve learned so far?
- How can I get my data into a wider format?
Objectives
- To be able to combine the different functions we have covered in tandem to create seamless chains of data handling
- Creating custom, complex data summaries
- Creating complex plots with grids of subplots
Motivation
This session is going to be a little different than the others. We will be working with more challenges and exploring different way of combining the things we have learned these days.
Before the break, and a little scattered through the sessions, we have been combining the things we have learned. It’s when we start using the tidyverse as a whole, all functions together that they start really becoming powerful. In this last session, we will be working on the things we have learned and applying them together in ways that uncover some of the cool things we can get done.
Lets say we want to summarise all the measurement variables, i.e. all the columns containing “_“. We’ve learned about summaries and grouped summaries. Can you think of a way we can do that using the things we’ve learned?
R
penguins |>
pivot_longer(contains("_"))
OUTPUT
# A tibble: 1,376 × 6
species island sex year name value
<fct> <fct> <fct> <int> <chr> <dbl>
1 Adelie Torgersen male 2007 bill_length_mm 39.1
2 Adelie Torgersen male 2007 bill_depth_mm 18.7
3 Adelie Torgersen male 2007 flipper_length_mm 181
4 Adelie Torgersen male 2007 body_mass_g 3750
5 Adelie Torgersen female 2007 bill_length_mm 39.5
6 Adelie Torgersen female 2007 bill_depth_mm 17.4
7 Adelie Torgersen female 2007 flipper_length_mm 186
8 Adelie Torgersen female 2007 body_mass_g 3800
9 Adelie Torgersen female 2007 bill_length_mm 40.3
10 Adelie Torgersen female 2007 bill_depth_mm 18
# ℹ 1,366 more rowsWe’ve done this before, why is it a clue now? Now that we have learned grouping and summarising, what if we now also group by the new name column to get summaries for each column as a row already here!
R
penguins |>
pivot_longer(contains("_")) |>
group_by(name) |>
summarise(mean = mean(value, na.rm = TRUE))
OUTPUT
# A tibble: 4 × 2
name mean
<chr> <dbl>
1 bill_depth_mm 17.2
2 bill_length_mm 43.9
3 body_mass_g 4202.
4 flipper_length_mm 201. Now we are talking! Now we have the mean of each of our observational columns! Lets add other common summary statistics.
R
penguins |>
pivot_longer(contains("_")) |>
group_by(name) |>
summarise(
mean = mean(value, na.rm = TRUE),
sd = sd(value, na.rm = TRUE),
min = min(value, na.rm = TRUE),
max = max(value, na.rm = TRUE)
)
OUTPUT
# A tibble: 4 × 5
name mean sd min max
<chr> <dbl> <dbl> <dbl> <dbl>
1 bill_depth_mm 17.2 1.97 13.1 21.5
2 bill_length_mm 43.9 5.46 32.1 59.6
3 body_mass_g 4202. 802. 2700 6300
4 flipper_length_mm 201. 14.1 172 231 That’s a pretty neat table! The repetition of
na.rm = TRUE in all is a little tedious, though. Let us use
an extra argument in the pivot longer to remove NAs in the
value column
R
penguins |>
pivot_longer(contains("_")) |>
drop_na(value) |>
group_by(name) |>
summarise(
mean = mean(value),
sd = sd(value),
min = min(value),
max = max(value)
)
OUTPUT
# A tibble: 4 × 5
name mean sd min max
<chr> <dbl> <dbl> <dbl> <dbl>
1 bill_depth_mm 17.2 1.97 13.1 21.5
2 bill_length_mm 43.9 5.46 32.1 59.6
3 body_mass_g 4202. 802. 2700 6300
4 flipper_length_mm 201. 14.1 172 231 Now we have a pretty decent summary table of our data.
Challenge 1
In our code making the summary table. Add another summary column for
the number of records, giving it the name n.
Try the n() function.
R
penguins |>
pivot_longer(contains("_")) |>
drop_na(value) |>
group_by(name) |>
summarise(
mean = mean(value),
sd = sd(value),
min = min(value),
max = max(value),
n = n()
)
OUTPUT
# A tibble: 4 × 6
name mean sd min max n
<chr> <dbl> <dbl> <dbl> <dbl> <int>
1 bill_depth_mm 17.2 1.97 13.1 21.5 342
2 bill_length_mm 43.9 5.46 32.1 59.6 342
3 body_mass_g 4202. 802. 2700 6300 342
4 flipper_length_mm 201. 14.1 172 231 342Challenge 2
Try grouping by more variables, like species and island, is the output what you would expect it to be?
R
penguins |>
pivot_longer(contains("_")) |>
drop_na(value) |>
group_by(name, species, island) |>
summarise(
mean = mean(value),
sd = sd(value),
min = min(value),
max = max(value),
n = n()
)
OUTPUT
`summarise()` has grouped output by 'name', 'species'. You can override using
the `.groups` argument.OUTPUT
# A tibble: 20 × 8
# Groups: name, species [12]
name species island mean sd min max n
<chr> <fct> <fct> <dbl> <dbl> <dbl> <dbl> <int>
1 bill_depth_mm Adelie Biscoe 18.4 1.19 16 21.1 44
2 bill_depth_mm Adelie Dream 18.3 1.13 15.5 21.2 56
3 bill_depth_mm Adelie Torgersen 18.4 1.34 15.9 21.5 51
4 bill_depth_mm Chinstrap Dream 18.4 1.14 16.4 20.8 68
5 bill_depth_mm Gentoo Biscoe 15.0 0.981 13.1 17.3 123
6 bill_length_mm Adelie Biscoe 39.0 2.48 34.5 45.6 44
7 bill_length_mm Adelie Dream 38.5 2.47 32.1 44.1 56
8 bill_length_mm Adelie Torgersen 39.0 3.03 33.5 46 51
9 bill_length_mm Chinstrap Dream 48.8 3.34 40.9 58 68
10 bill_length_mm Gentoo Biscoe 47.5 3.08 40.9 59.6 123
11 body_mass_g Adelie Biscoe 3710. 488. 2850 4775 44
12 body_mass_g Adelie Dream 3688. 455. 2900 4650 56
13 body_mass_g Adelie Torgersen 3706. 445. 2900 4700 51
14 body_mass_g Chinstrap Dream 3733. 384. 2700 4800 68
15 body_mass_g Gentoo Biscoe 5076. 504. 3950 6300 123
16 flipper_length_mm Adelie Biscoe 189. 6.73 172 203 44
17 flipper_length_mm Adelie Dream 190. 6.59 178 208 56
18 flipper_length_mm Adelie Torgersen 191. 6.23 176 210 51
19 flipper_length_mm Chinstrap Dream 196. 7.13 178 212 68
20 flipper_length_mm Gentoo Biscoe 217. 6.48 203 231 123Challenge 3
Create another summary table, with the same descriptive statistics (mean, sd ,min,max and n), but for all numerical variables. Grouped only by the variable names.
R
penguins |>
pivot_longer(where(is.numeric)) |>
drop_na(value) |>
group_by(name) |>
summarise(
mean = mean(value),
sd = sd(value),
min = min(value),
max = max(value),
n = n()
)
OUTPUT
# A tibble: 5 × 6
name mean sd min max n
<chr> <dbl> <dbl> <dbl> <dbl> <int>
1 bill_depth_mm 17.2 1.97 13.1 21.5 342
2 bill_length_mm 43.9 5.46 32.1 59.6 342
3 body_mass_g 4202. 802. 2700 6300 342
4 flipper_length_mm 201. 14.1 172 231 342
5 year 2008. 0.818 2007 2009 344Plotting summaries
Now that we have the summaries, we can use them in plots too! But keep typing or copying the same code over and over is tedious. So let us save the summary in its own object, and keep using that.
R
penguins_sum <- penguins |>
pivot_longer(contains("_")) |>
drop_na(value) |>
group_by(name, species, island) |>
summarise(
mean = mean(value),
sd = sd(value),
min = min(value),
max = max(value),
n = n()
) |>
ungroup()
OUTPUT
`summarise()` has grouped output by 'name', 'species'. You can override using
the `.groups` argument.We can for instance make a bar chart with the values from the summary statistics.
R
penguins_sum |>
ggplot(aes(x = island,
y = mean,
colour = species)) +
geom_point() +
facet_wrap(~ name, scales = "free_y")
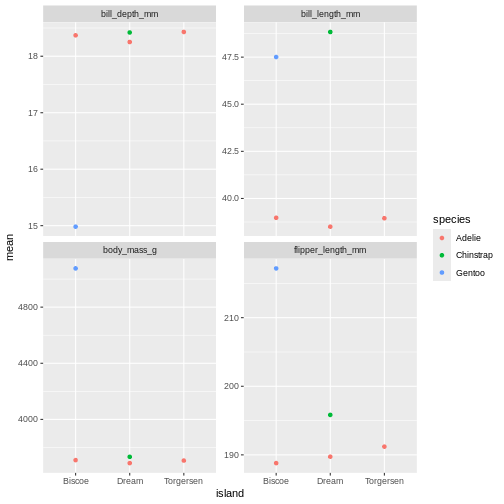
oh, but the points are stacking on top of each other and are hard to see. T
R
penguins_sum |>
ggplot(aes(x = island,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
facet_wrap(~ name, scales = "free_y")
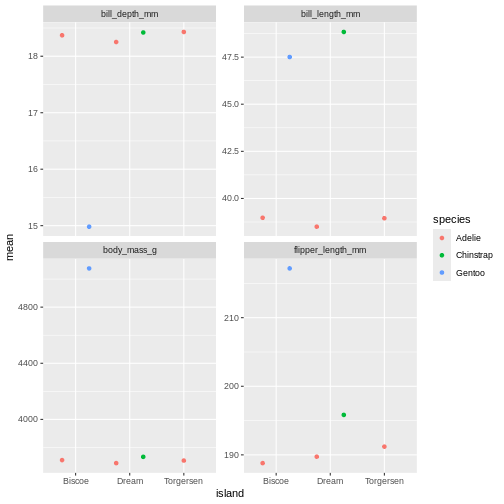
That is starting to look like something nice. What position_dodge is doing, is move the dts to each side a little, so they are not directly on top of each other, but you can still see them and which island they belong to clearly.
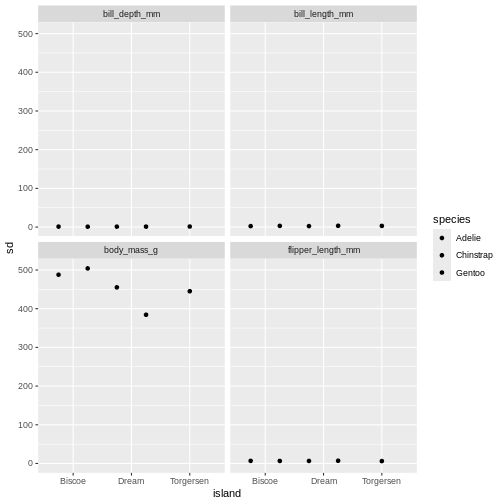
Challenge 4
Create a point plot based om the penguins summary data, where the standard deviations are on the y axis and species are on the x axis. Make sure to dodge the bar for easier comparisons. Create subplots on the different observational types
Use facet_wrap()
R
penguins_sum |>
ggplot(aes(x = island,
y = sd,
fill = species)) +
geom_point(position = position_dodge(width = 1)) +
facet_wrap(~ name)
Challenge 5
Change it so that species is both on the x-axis and the colour for
the bar chart, and remove the dodge. What argument do you need to add to
facet_wrap() to make the y-axis scale vary freely between
the subplots? Why is this plot misleading?
R
penguins_sum |>
ggplot(aes(x = species,
y = sd,
fill = species)) +
geom_point(position = position_dodge(width = 1)) +
facet_wrap(~ name, scales = "free")
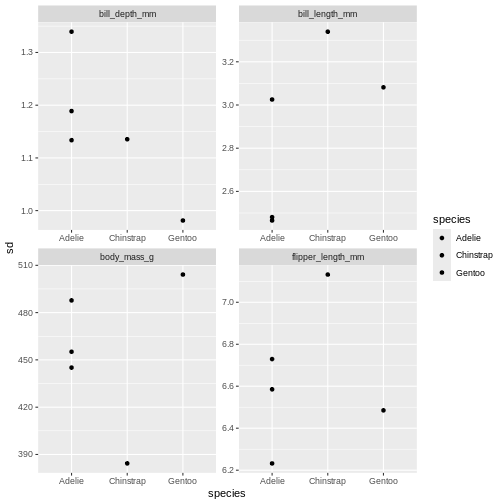 The last plot is misleading because the data we have summary data by
species and island. Ignoring the island in the plot, means that the
values for the different measurements cannot be distinguished from
eachother.
The last plot is misleading because the data we have summary data by
species and island. Ignoring the island in the plot, means that the
values for the different measurements cannot be distinguished from
eachother.
A common thing to add to this type of plot, is the confidence intervals, or the error bars. This is calculated by the standard error, which we dont have, but for the sake of showing how to add error bars, we will use the standard deviation in stead.
To do that, we add the geom_errorbar() function to the
ggplot calls. geom_errorbar is a little different than
other geoms we have seen, it takes very specific arguments, namely the
minimum and maximum value the error bars should span. In our case, it
would be the mean - sd, for minimum, and the mean + sd for the
maximum.
R
penguins_sum |>
ggplot(aes(x = island,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean + sd
)) +
facet_wrap(~ name, scales = "free_y")
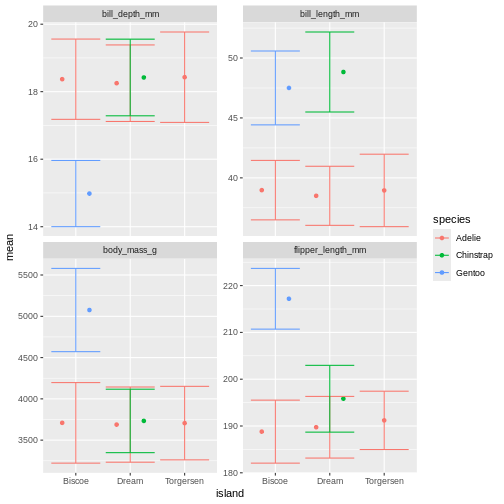
Right, so now we have error bars, but they dont connect to the dots! Perhaps we can dodge those too?
R
penguins_sum |>
ggplot(aes(x = island,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean + sd
),
position = position_dodge(width = 1)) +
facet_wrap(~ name, scales = "free_y")
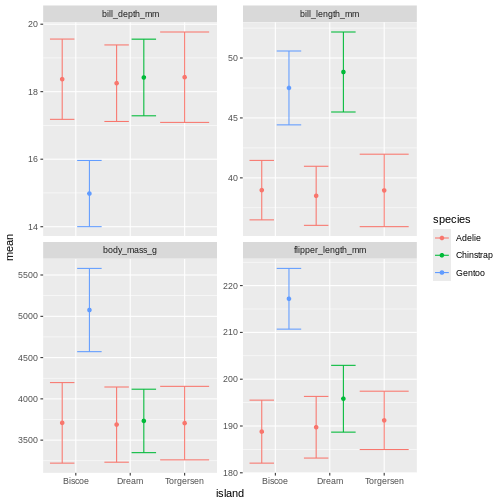
Challenge 6
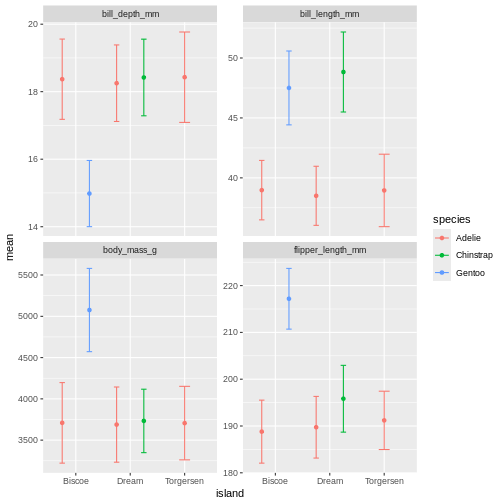
The width of the top horizontal lines in the error bars are are little too wide. Try adjusting them by setting the width argument to 0.3
R
penguins_sum |>
ggplot(aes(x = island,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean + sd
),
position = position_dodge(width = 1),
width = .2) +
facet_wrap(~ name, scales = "free_y")
Facetting as a grid
But we can get even more creative! Lets recreate our summary table, and add year as a grouping, so we can get an idea of how the measurements change over time.
R
penguins_sum <- penguins |>
pivot_longer(contains("_")) |>
drop_na(value) |>
group_by(name, species, island, year) |>
summarise(
mean = mean(value),
sd = sd(value),
min = min(value),
max = max(value),
n = n()
) |>
ungroup()
OUTPUT
`summarise()` has grouped output by 'name', 'species', 'island'. You can
override using the `.groups` argument.R
penguins_sum
OUTPUT
# A tibble: 60 × 9
name species island year mean sd min max n
<chr> <fct> <fct> <int> <dbl> <dbl> <dbl> <dbl> <int>
1 bill_depth_mm Adelie Biscoe 2007 18.4 0.585 17.2 19.2 10
2 bill_depth_mm Adelie Biscoe 2008 18.1 1.20 16.2 21.1 18
3 bill_depth_mm Adelie Biscoe 2009 18.6 1.44 16 20.7 16
4 bill_depth_mm Adelie Dream 2007 18.7 1.21 16.7 21.2 20
5 bill_depth_mm Adelie Dream 2008 18.3 0.993 16.1 20.3 16
6 bill_depth_mm Adelie Dream 2009 17.7 0.994 15.5 20.1 20
7 bill_depth_mm Adelie Torgersen 2007 19.0 1.47 17.1 21.5 19
8 bill_depth_mm Adelie Torgersen 2008 18.1 1.11 16.1 19.4 16
9 bill_depth_mm Adelie Torgersen 2009 18.0 1.20 15.9 20.5 16
10 bill_depth_mm Chinstrap Dream 2007 18.5 1.00 16.6 20.3 26
# ℹ 50 more rowsAnd then let us re-create our last plot with this new summary table.
R
penguins_sum |>
ggplot(aes(x = island,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean + sd
),
width = 0.3,
position = position_dodge(width = 1)) +
facet_wrap(~ name, scales = "free_y")
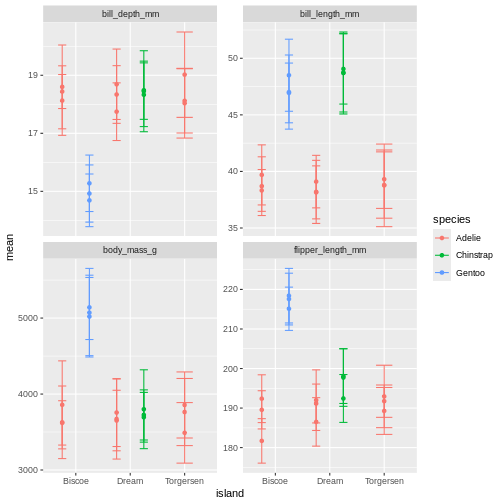
What is happening here? Because we’ve now added year to the groups in the summary, we have multiple means per species and island, for each of the measurement years. So we need to add something to the plot so we can tease those appart. We have added to variables to the facet before. Remember how we did that?
Challenge 7
The width of the top horizontal lines in the error bars are are little too wide. Try adjusting them by setting the width argument to 0.3
R
penguins_sum |>
ggplot(aes(x = island,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean + sd
),
position = position_dodge(width = 1),
width = .2) +
facet_wrap(~ name + year, scales = "free_y")
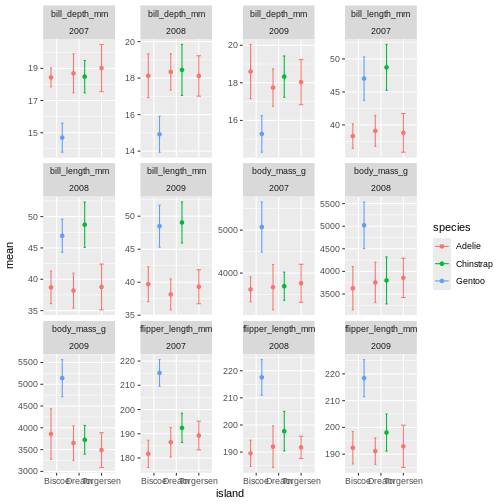
OK, so now we have it all. But its a little messy to compare over time, and what are we really looking at? I find it often makes more sense to plot time variables on the x-axis, and then facets over categories. Lets switch that up.
R
penguins_sum |>
ggplot(aes(x = year,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean + sd
),
position = position_dodge(width = 1),
width = .2) +
facet_wrap(~ name + island, scales = "free_y")
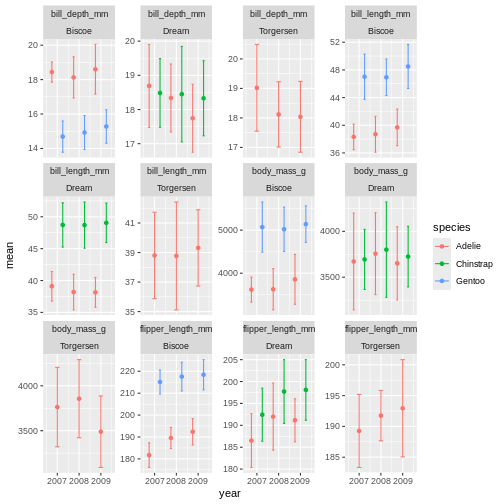 ok, so we got what we asked, the year part makes more sense, but its a
very “busy” plot. Its really quite hard to compare everything from
Bisoe, or all the Adelie’s, to each other. How can we make it
easier?
ok, so we got what we asked, the year part makes more sense, but its a
very “busy” plot. Its really quite hard to compare everything from
Bisoe, or all the Adelie’s, to each other. How can we make it
easier?
We will switch facet_wrap() to facet_grid()
which creates a grid of subplots. The formula for the grid is using both
side of the ~ sign. And you can think of it like
rows ~ columns. So here we are saying we want the
island values as rows, and name values as
columns in the plot grid.
R
penguins_sum |>
ggplot(aes(x = year,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean + sd
),
position = position_dodge(width = 1),
width = .2) +
facet_grid(island ~ name)
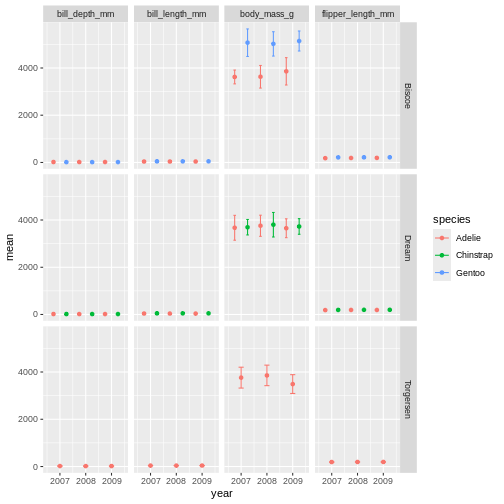
Challenge 8
It is hard to see the different metrics in the subplots, because they are all on such different scales. Try setting the y-axis to be set freely to allow differences betweem the subplots. Was this the effect you expected?
R
penguins_sum |>
ggplot(aes(x = year,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean + sd
),
position = position_dodge(width = 1),
width = .2) +
facet_grid(island ~ name, scales = "free_y")
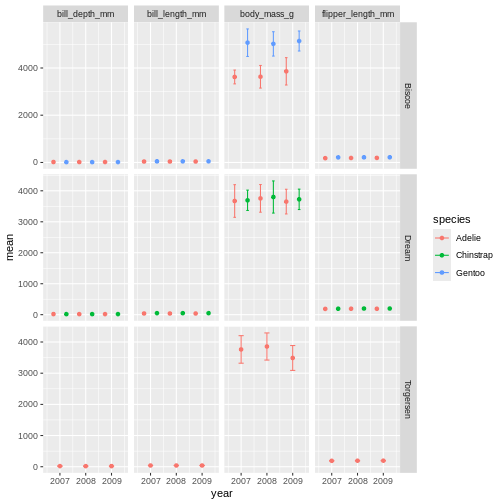
Challenge 9
Try switching up what is plotted as rows and columns in the facet. Does this help the plot?
R
penguins_sum |>
ggplot(aes(x = year,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean + sd
),
position = position_dodge(width = 1),
width = .2) +
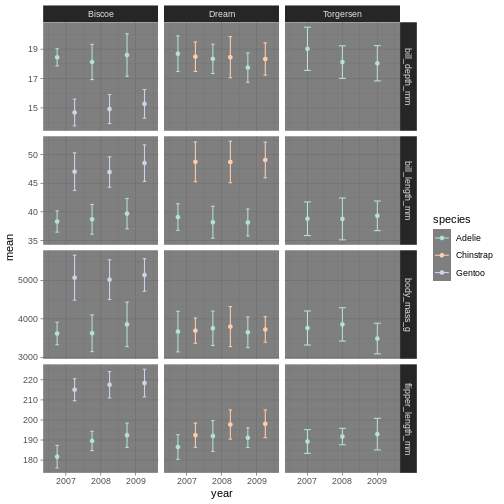
facet_grid(name ~ island, scales = "free_y")
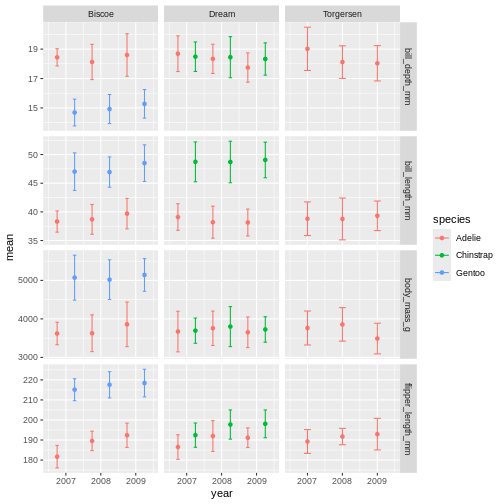
facet_grid is more complex than facet_wrap as
it will always force the y-axis for rows, and x-axis for columns remain
the same. So wile setting scales to free will help a little, it will
only do so within each row and column, not each subplot. When the
results do not look as you like, swapping what are rows and columns in
the grid can often create better results.
Altering ggplot colours and theme
We now have a plot that is quite nicely summarising the data we have. But we want to customise it more. While the defaults in ggplot are fine enough, we usually want to improve it from the default look.
Before we do that, lets save the plot as an object, so we dont have to keep track of the part of the code we are not changing. Saving a ggplot object is just like saving a dataset object. We have to assign it a name at the beginning.
R
penguins_plot <- penguins_sum |>
ggplot(aes(x = year,
y = mean,
colour = species)) +
geom_point(position = position_dodge(width = 1)) +
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean + sd
),
position = position_dodge(width = 1),
width = .2) +
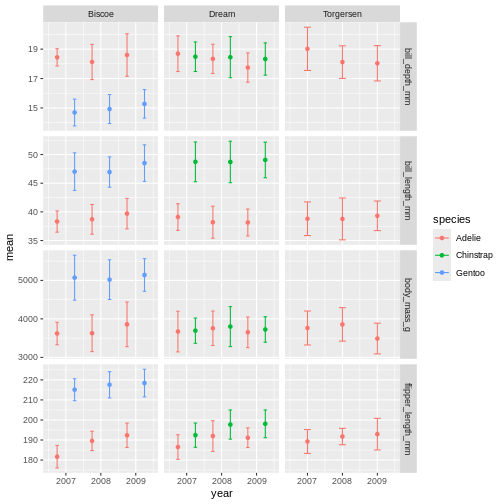
facet_grid(name ~ island, scales = "free_y")
Did you notice that it did not make a new plot? Just like when you assign a data set it wont show in the console, when you assign a plot, it wont show in the plot pane.
To re-initiate the plot in the plot pane, write its name in the console and press enter.
R
penguins_plot
From there, we can keep adding more ggplot geoms or facets etc. In this first version, we will add a “theme”. A theme is a change of the overall “look” of the plot.
R
penguins_plot +
theme_classic()
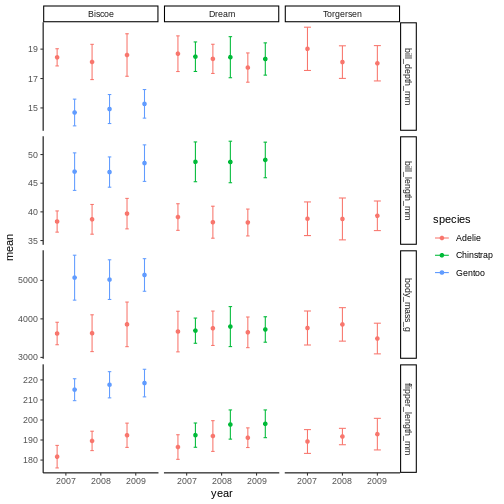 the classic theme is preferred by many journals, but for facet grid, its
not super nice, since we loose grid information.
the classic theme is preferred by many journals, but for facet grid, its
not super nice, since we loose grid information.
R
penguins_plot +
theme_light()
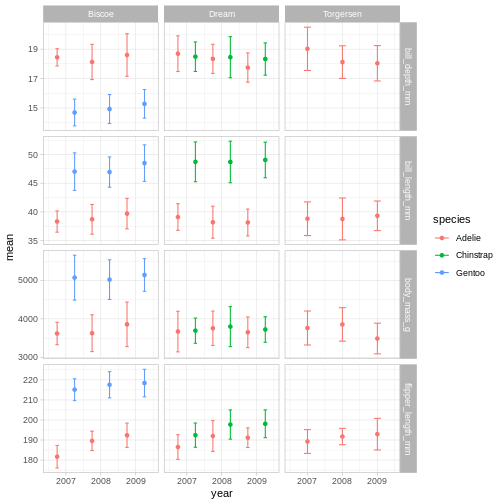 Theme light could be a nice option, but the white text of light grey
makes the panel text hard to read.
Theme light could be a nice option, but the white text of light grey
makes the panel text hard to read.
R
penguins_plot +
theme_dark()
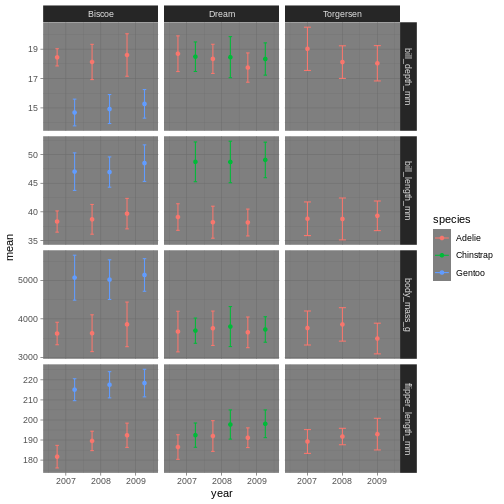
Theme dark could theoretically be really nice, but then we’ll need other colours for the points and error bars!
Challenge 10
Try different themes and find one you like.
You can type “theme” and press the tab button, to look at all the possibilities.
What themes did you find that you liked?
We are going to have a go at theme_linedraw which has a
simple but clear design.
R
penguins_plot +
theme_linedraw()
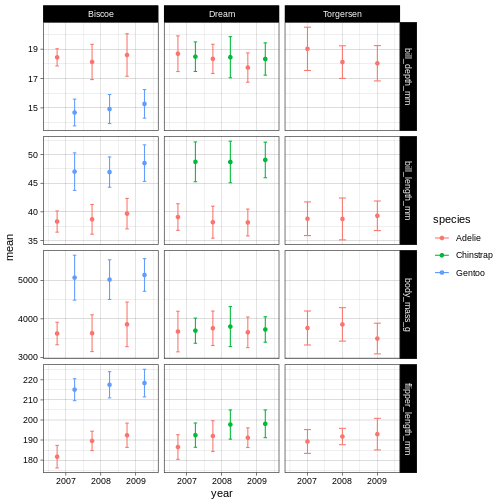
Now that we have a theme, we can have a look at changing the colours of the points and error bars. We do this through something called “scales”.
R
penguins_plot +
theme_linedraw() +
scale_colour_brewer(palette = "Dark2")
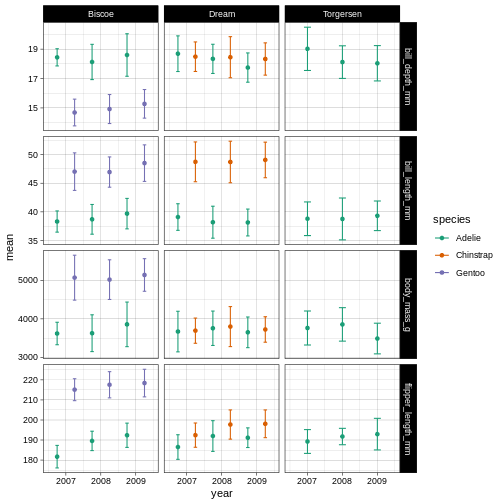
So here, we are changing the colour aesthetic, using a “brewer” palette “Dark2”. What is a brewer palette? THe brewer palettes are a curated library of colour palettes to choose from in ggplot. You can have a peak at all possible brewer palettes by typing
R
RColorBrewer::display.brewer.all()

Challenge 11
Try another brewer palette by replacing the palette name with another in the brewer list of palettes.
R
penguins_plot +
theme_linedraw() +
scale_colour_brewer(palette = "Accent")
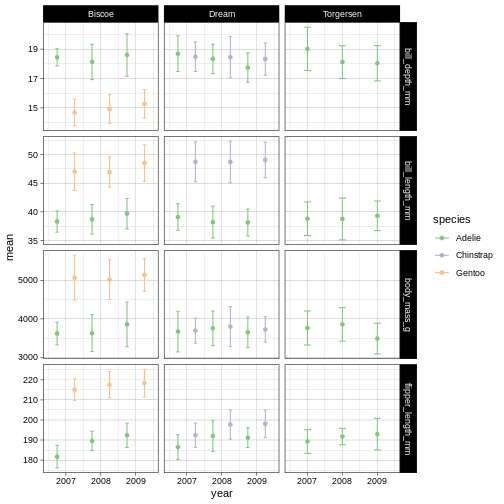
Challenge 12
Apply the dark theme in stead, and a pastel colour palette.
R
penguins_plot +
theme_dark() +
scale_colour_brewer(palette = "Pastel2")
Amazing! We have now adapted our plot to look nicer and more to our liking. There are plenty of packages out there with specialised themes and colour palettes to choose from. Harry Potter colours, Wes Anderson colours, Ghibli move colours. You can find almost anything you like!
