Evolutionary Genome Mining
Overview
Teaching: 20 min
Exercises: 20 minQuestions
What is Evolutionary Genome Mining?
Which kind of BGCs can EvoMining find?
What do I need in order to run an evolutionary genome mining analysis?
Objectives
Understand EvoMining pipeline.
Run an example of evolutionary analysis in cpsG gene family.
Explore MicroReact interactive output interface.
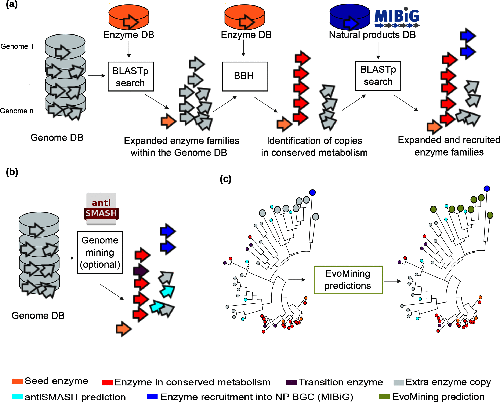
Usually, bioinformatics tools related to the prediction of Natural Products (NP) biosynthetic genes try to find metabolic pathways of enzymes that are known to be related with the synthesis of secondary metabolites. However, these approaches fail for the discovery of novel biosynthetic systems. Thus, EvoMining tries to circumvent this problem by detecting novel enzymes that may be implicated in the synthesis of new natural products in Bacteria.
To know more about EvoMining you can read Selem et al, Microbial Genomics 2019.
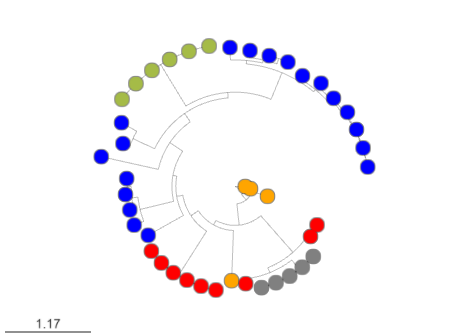
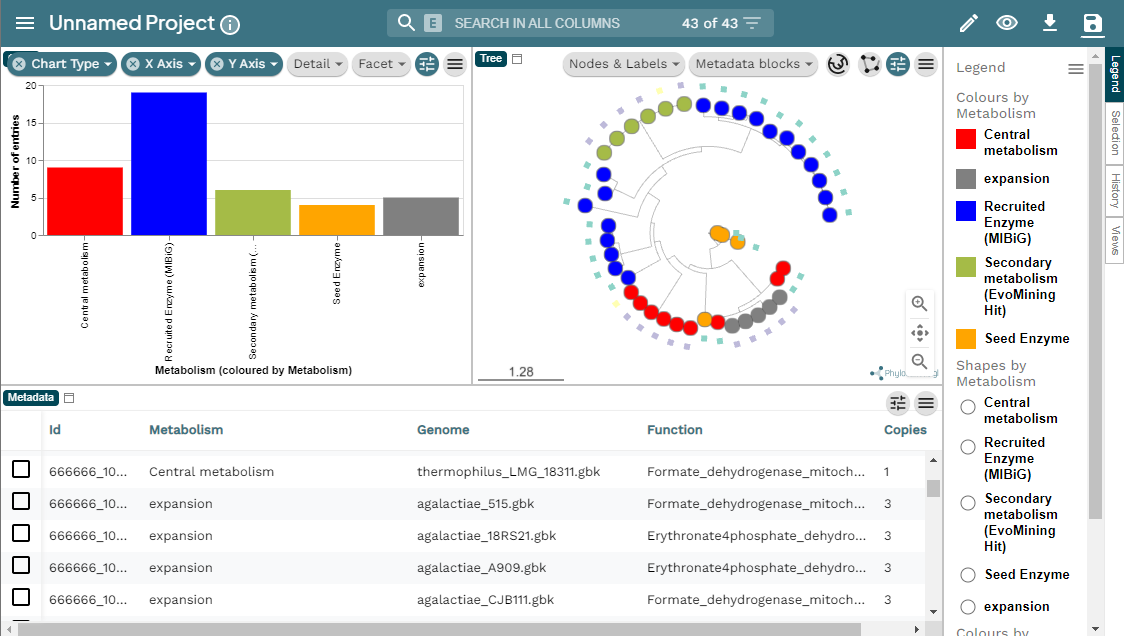
EvoMining searches protein expansions that may have evolved from the conserved metabolism into a specialized metabolism. It builds phylogenetic trees based on all the protein copies of a certain enzyme in a given genome database. The output tree differentiates copies that are related with the conserved metabolism, copies that are known to be implicated in discovered NP-producing-BGCs i.e. BGCs from MiBIG database and, optionally, protein copies that belong to BGCs predicted by antiSMASH. Finally, some branch in the tree will be depicted as “EvoMining hits”, which represent enzyme expansions that are evolutionary closer to those copies related with the secondary metabolism (MiBIG or antiSMASH BGCs) than to those related with the conserved (primary) metabolism.
Run evomining image
First, place yourself at your working directory.
$ cd ~/pan_workshop/results/genome-mining/corason-conda/EXAMPLE2
$ ls
CORASON_GENOMES Corason_Rast.IDs cpsg.query GENOMES output
The general structure of a docker container is shown in the next bash-box. Note that
it requires to specify which docker container will run. Optionally,
with -v flag it is possible to share a directory with the container,
with -p flag a port is shared and it is also possible to specify which
program will run inside the container.
$ docker run --rm -i -t -v <your local directory>:<inside docker directory> -p <inside port>:80 <docker container> <program inside docker>
EvoMining is inside a docker container, so the general structure to start your analysis will be as follows:
$ docker run --rm -i -t -v $(pwd):/var/www/html/EvoMining/exchange -p 8080:80 nselem/evomining:latest /bin/bash
Let’s explain the pieces of this line.
| command | Explanation |
|---|---|
| docker | tells the system that we are running a docker command |
| run | the command that we are running is to run a docker container |
| –rm | this container will be removed after closed |
| -i | this container allows user interaction |
| -t | this interaction will be through a terminal |
| -v | a data volume (directory) will be shared between your local machine and the container |
| -p | a port will allow a web based app |
However, sometimes the port 80 is busy, in that case you can
use other ports like 8080 or 8084. If this is the case, please use the port 80X
where X is a number between 01..30 provided by your instructor.
$ docker run --rm -i -t -v $(pwd):/var/www/html/EvoMining/exchange -p 8080:80 nselem/evomining:latest /bin/bash
$ docker run --rm -i -t -v $(pwd):/var/www/html/EvoMining/exchange -p 8084:80 nselem/evomining:latest /bin/bash
If your docker container worked, now you will see in your terminal
a new prompt. Instead of the usual dollar sign, there should be a number
# at the beginning of your terminal. This is because now you are inside
the docker container and you have sudo permissions inside the docker.
#
To exit container use exit
# exit
And now your prompt must be back in the dollar sign
$
Set EvoMining genomic database
Start the container again with your corresponding port.
$ docker run --rm -i -t -v $(pwd):/var/www/html/EvoMining/exchange -p 80X:80 nselem/evomining:latest /bin/bash
Though we will NOT run the test EvoMining command, it looks as follows:
# perl startEvoMining.pl
Instead of that, customize the genomic database by using the same as CORASON.
Notice that EvoMining requires RAST-like annotated genomes and for this reason
we are using the fasta files that CORASON converts from our gbk inputs.
# perl startEvoMining.pl -g GENOMES -r Corason_Rast.IDs
Finally, remember that X means your user-number and open your browser
at the address: http://132.248.196.38:80X/EvoMining/html/index.html. Once there,
just click the start button and enjoy! (click on the submit buttons!)
When you finish using this container, please exit it.
#exit
Visualize your results in MicroReact
Firstly, you have to run all the pipeline in the website:
http://
EvoMining outputs are stored in the directory <conserved-db>_<natural-db>_<genomes-db>
$ ls
To explore EvoMining outputs, you need to upload 1.nwk and 1.csv files to microReact. There are many methods to download files from the server to your local computer.
If you are using JupyterHub you explore the file folders and select the files and then press the download button.
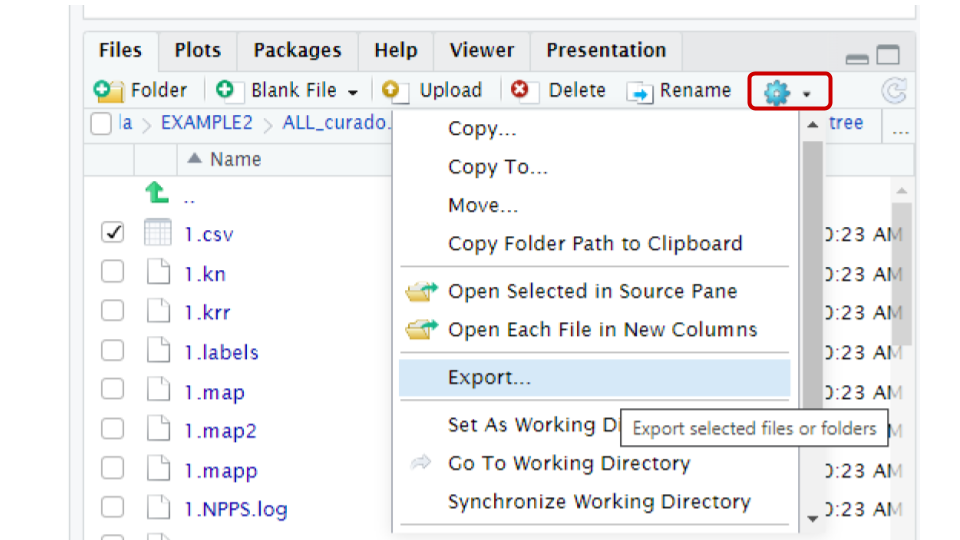
You can use the export button in the file panel of R studio.
To download the files, first in the files panel open in
the directory ~/pan_workshop/results/genome-mining/corason-conda/EXAMPLE2/ALL_curado.fasta_MiBIG_DB.faa_GENOMES/blast/seqf/tree,
Then, select files 1.nwk and 1.csv in that directory, click more
in the engine icon, and select the export option in the menu. The files
will be downloaded to your local computer, and now you will be able to
upload them to MicroReact.
Alternatively, If your prefer to use the terminal to download files
the scp protocol can download the files into your local machine.
scp betterlab@132.248.196.38:~/pan_workshop/results/genome-mining/corason-conda/EXAMPLE2/ALL_curado.fasta_MiBIG_DB.faa_GENOMES/blast/seqf/tree/1.nwk ~/Downloads/.
scp betterlab@132.248.196.38:~/pan_workshop/results/genome-mining/corason-conda/EXAMPLE2/ALL_curado.fasta_MiBIG_DB.faa_GENOMES/blast/seqf/tree/1.csv ~/Downloads
Here you can find the MicroReact visualization of this EvoMining run.
Other resources
To run EvoMining with a larger conserved-metabolite DB you can use EvoMining Zenodo data.
To explore more EvoMining options, please explore EvoMining wiki.
Set the conserved-enzymes database
When using EvoMining, oftenly you will desire to construct your own conserved enzymes database. To know more about how to configure a database, consult the EvoMining wiki in the EvoMining databases part. Natural products database could also be replaced for another set of genes that are “true positives”, for example a set of regulatory genes.
As an example, transform the file cpsg.query into the format of this database.
This file contains the aminoacid sequence of the cpsG gene. Firstly, copy this file
into what will become the conserved-enzymes database.
$ cp cpsg.query cpsg_cdb
Now, it requires some editing. Open nano editor and change the first line >cpsg to
>SYSTEM1|1|phosphomannomutase|Saga . EvoMining conserved-database needs a
four-field format pipe-separated that contains; the name of the metabolic system to which the
enzyme belongs (SYSTEM1), a consecutive number of the enzyme (1 in this case),
the function of the enzyme, and finally, an abbreviation of the organism Saga, (S. Agalactiae).
The reason behind this is that this was the way we needed EvoMining for its first use and we have not changed the headers since.
$ nano cpsg_cdb
>SYSTEM1|1|phosphomannomutase|Saga
MIFVTVGTHEQQFNRLIKEVDRLKGTGAIDQEVFIQTGYSDFEPQNCQWSKFLSYDDMNSYMKEAEIVITHGGPATFMSVISLGKLPVVVPRRKQFGEHINDHQIQFLKKIAHLYPLAWIED
Run your EvoMining docker
$ docker run --rm -i -t -v $(pwd):/var/www/html/EvoMining/exchange -p 80X:80 nselem/evomining:latest /bin/bash
and inside this new container:
# perl startEvoMining.pl -g GENOMES -r Corason_Rast.IDs -c cpsg_cdb
Use the website again and think about the results.
Exercise 1. Set EvoMining parameters
Complete the blanks in the following EvoMining run:
actinoSMASHA file with the ids of antiSMASH recognized genes.Actinosa directory with RAST-like fasta and annotation files.Histidine-dbA fasta file with some proteins in the histidine pathway.
Actinos.idstabular files with the RAST ids and the name of the organisms.# perl starEvoMining.pl -g ____ -c _____ -r _____ -a ___________Solution
# perl starEvoMining.pl -g Actinos -c Histidine-db -r Actinos.ids -a actinoSMASHActinos is the genomic database, Histidine-db is the conserved-enzymes database, Actinos.ids is the file that relates Rast ids with the organisms names, and actinoSMASH contains the genes identified by antiSMASH.
Discussion 1. Retro EvoMining in enzyme database
What do you learn from running in a conserved-enzymes database the gene cpsG that is part of a specialized BGC?
Solution
cpsG does not have extra copies in Streptococcus agalactiae, so there are no expansions that may be functional divergent. cpsG single copies in the genomes look red-colored in EvoMining output, as if they belong to the conserved-metabolism. However, this is not the case, the color is because there is only one copy and it is merged into MIBiG true-positives because it was originally a gene in the specialized metabolism. So it is important to know the seed enzymes.
cpsg_cdb_MiBIG_DB.faa_GENOMES
ARTS is another evolutionary genome mining software with its corresponding database ARTS-db .
Evolutionary Genome Mining
To learn more about Genome Mining you can read these references:
- The confluence of big data and evolutionary genome mining for the discovery of natural products. Chevrette et al, 2021`
- Evolutionary Genome Mining for the Discovery and Engineering of Natural Product Biosynthesis. Chevrette et al, 2022
More about docker
To see the running container use
ps$ docker ps
If there are containers in use you will see a list of all of them.
2f879ba6e337 nselem/evomining:latest "/bin/bash" 11 hours ago Up 11 hours 0.0.0.0:8014->80/tcp, :::8014->80/tcp relaxed_dirac
To stop the running container use
docker stopand to remove them usedocker rm$ docker stop relaxed_dirac $ docker remove relaxed_dirac2f879ba6e337can be downloaded by specifying our queries with
--format genbank.
Key Points
EvoMining is a command-line tool that performs evolutionary genome mining over gene families
EvoMining hits can belong to new BGC
MicroReact is an interactive genomic visualizer compatible with EvoMining output