Finding Variation on Genomic Vicinities
Overview
Teaching: 20 min
Exercises: 15 minQuestions
How can I follow variation in genomic vicinities given a reference BGC?
Which gene families are the conserved part of a BGC family?
Objectives
Learn to find BGC-families
Visualize variation in a genomic vicinity
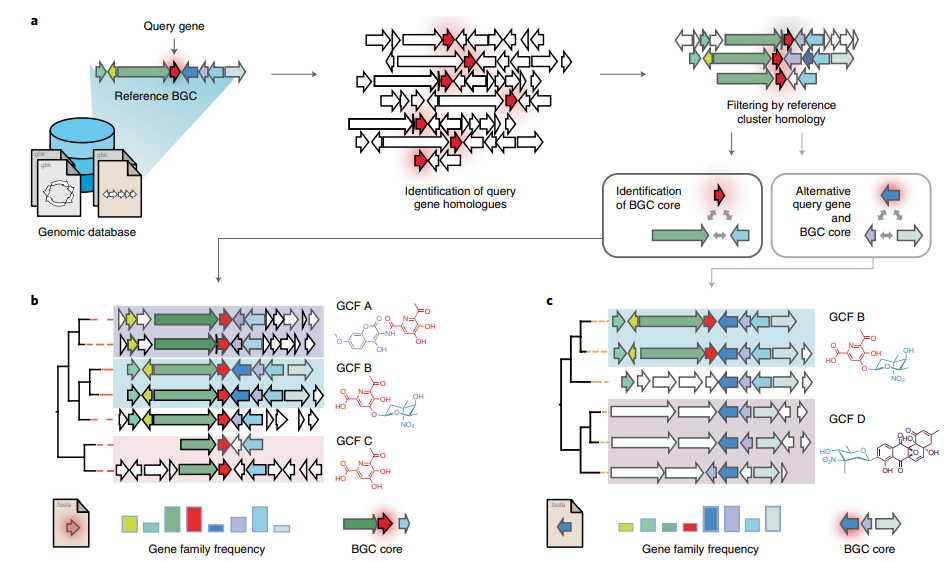
CORASON is a visual tool that identifies gene clusters that share a common genomic core and reconstructs multi-locus phylogenies of these gene clusters to explore their evolutionary relationships. CORASON was developed to find and prioritize biosynthetic gene clusters (BGC), but can be used for any kind of clusters.
Input: query gene, reference BGC and genome database. Output: SVG figure with BGC in the family sorted according to the multi-locus phylogeny of the core genes.
Advantages
- SVG graphs Scalable graphs that allow metadata easy display.
- Interactive, CORASON is not a static database, it allows to explore your own genomes.
- Reproducibility, since CORASON runs on docker and conda, the containerization allows to always perform the same analysis even if you change your Linux/perl/blast/muscle/Gblocks/quicktree local distributions.
CORASON conda
Here we are testing the new stand-alone corason in a conda environment with gbk as input-files. Firstly, activate the corason-conda environment.
$ conda deactivate
$ conda activate /miniconda3/envs/corason
(corason) user@server:~$
With the environment activated, all CORASON-dependencies are ready to be used. The next step is to clone CORASON-software from its GitHub repository. Firstly, place yourself at the results directory and then clone CORASON-code.
$ mkdir -p ~/pan_workshop/results/genome-mining
$ cd ~/pan_workshop/results/genome-mining
$ git clone https://github.com/miguel-mx/corason-conda.git
$ ls
The GitHub CORASON repository must be now in your directory.
corason-conda
Change directory by descending to EXAMPLE2. The file cpsg.query,
contains the reference protein cpsG, whose encoding gene
is part of the
polysaccharide BGC
produced by some S. agalactiae
$ cd corason-conda/EXAMPLE2
$ ls
cpsg.query
As genomic database we will use the prokka-annotated gbk files of S. agalactiae.
This database will be stored in the reserved directory CORASON_GENOMES.
$ mkdir CORASON_GENOMES
$ cp ~/pan_workshop/results/annotated/Streptococcus_agalactiae_*/*gbk CORASON_GENOMES/
CORASON was written to be used with RAST annotation as
input files, in this case we are using a genome database
composed of .gbk files. Thus, we need to convert
gbk files into CORASON-compatible input files.
$ ../CORASON/gbkIndex.pl CORASON_GENOMES ../CORASON CORASON_GENOMES
Directory CORASON_GENOMES
../CORASON/gbk_to_fasta.pl CORASON_GENOMES agalactiae_18RS21_prokka.gbk 100001 ../CORASON
../CORASON/gbk_to_fasta.pl CORASON_GENOMES agalactiae_515_prokka.gbk 100002 ../CORASON
../CORASON/gbk_to_fasta.pl CORASON_GENOMES agalactiae_A909_prokka.gbk 100003 ../CORASON
../CORASON/gbk_to_fasta.pl CORASON_GENOMES agalactiae_CJB111_prokka.gbk 100004 ../CORASON
../CORASON/gbk_to_fasta.pl CORASON_GENOMES agalactiae_COH1_prokka.gbk 100005 ../CORASON
../CORASON/gbk_to_fasta.pl CORASON_GENOMES agalactiae_H36B_prokka.gbk 100006 ../CORASON
../CORASON/gbk_to_fasta.pl CORASON_GENOMES thermophilus_LMD-9_prokka.gbk 100007 ../CORASON
../CORASON/gbk_to_fasta.pl CORASON_GENOMES thermophilus_LMG_18311_prokka.gbk 100008 ../CORASON
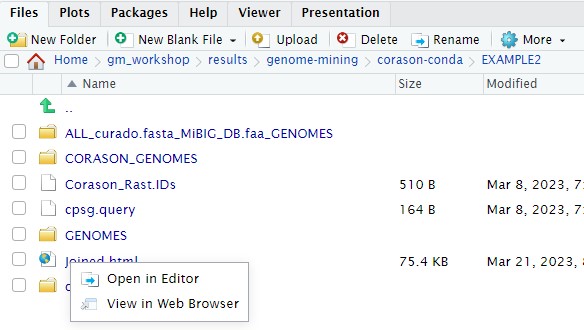
Now, all converted genome files, aminoacid fasta files (.faa) and annotation files (.txt) should be placed in the ‘GENOMES directory’ one level up outside output.
$ mv output/* .
$ ls
CORASON_GENOMES Corason_Rast.IDs cpsg.query GENOMES output
Finally, we have the query enzyme, the IDs file and a genomic database of S. agalactiae in the same directory. Run CORASON with cpsg.query as query with 1000006 as an example of reference BGC.
$ ../CORASON/corason.pl -q cpsg.query -s 100006 -rast_ids Corason_Rast.IDs
>> 100001_666.input Cluster1237 Cluster1245 Cluster1253 cpsg.query_BGC.tre cpsg.query_tree.svg
>> 100002_2002.input Cluster1238 Cluster1246 Cluster1254 cpsg.query.BLAST Frequency
>> 100003_1173.input Cluster1239 Cluster1247 Cluster1255 cpsg.query.BLAST.pre GBK
>> 100004_35.input Cluster1240 Cluster1248 Cluster1256 cpsg.query.parser Joined.svg
>> 100005_912.input Cluster1241 Cluster1249 Cluster1257 cpsg.query.parser.pre MINI
>> 100006_1247.input Cluster1242 Cluster1250 Concatenados.faa cpsg.query_PrincipalHits.tre PrincipalHits.muscle
>> 100007_1022.input Cluster1243 Cluster1251 CORASON cpsg.query_Report PrincipalHits.muscle-gb
>> 100008_1024.input Cluster1244 Cluster1252 cpsg.query cpsg.query.svg TempConcatenados.faa
Finally, we have all the genomic vicinities sorted phylogenetically according to the genes in the core-cluster. We can either see the output directly on the server or download the resulting svg file to our local computer. The format svg stands for scalable vector graphics, we are save it as an html file in order to get the option of open it as html. To see it in the server we must first copy the Joined.svg into Joined.html, then, go to the EXAMPLE2 directory in the r-studio files panel located at the bottom right. Once there clik in the Joined.html file and chose the option View in Web Browser. The corason figure will output in a new tab.
$ cp output/cpsg.query-output/Joined.svg Joined.html
You can also use scp in a local terminal to download the resulting svg file to your local computer.
$ scp (remoto)/corason-conda/EXAMPLE2/output/cpsg.query-output Downloads/.
CORASON has several installation and a conda beta versions
CORASON has one docker installation where you have to go inside the docker container, its input are RAST files. The script run_corason belongs to another installation that accepts gbk as input files. And finally, this is a beta version of CORASON conda container.
Key Points
CORASON is a command-line tool that finds BGC-families
Genomic vicinity variation is organized phylogenetically according to the conserved genes in the BGC-family