How Snakemake plans its jobs
Last updated on 2025-02-25 | Edit this page
Overview
Questions
- How do I visualise a Snakemake workflow?
- How does Snakemake avoid unecessary work?
- How do I control what steps will be run?
Objectives
- View the DAG for our pipeline
- Understand the logic Snakemake uses when running and re-running jobs
For reference, this is the Snakefile you should have to start the episode.
The DAG
You may have noticed that one of the messages Snakemake always prints is:
OUTPUT
Building DAG of jobs...A DAG is a Directed Acyclic Graph and it can be pictured like so:
The above DAG is based on our existing rules, and shows all the jobs Snakemake would run to trim, count and quantify the ref1 sample.
Note that:
- A rule can appear more than once, with different wildcards (a rule plus wildcard values defines a job)
- A rule (here, calculate_difference) may not be used at all, if it is not required for the target outputs
- The arrows show dependency ordering between jobs
- Snakemake can run the jobs in any order that doesn’t break dependency - for example kallisto_quant cannot run until both kallisto_index and trimreads have completed, but it may run before or after countreads
- This is a work list, not a flowchart, so there are no if/else decisions or loops - Snakemake runs every job in the DAG exactly once
- The DAG depends both on the Snakefile and on the requested target outputs, and the files already present
- When building the DAG, Snakemake does not look at the shell part of the rules at all - only when running the DAG will Snakemake check that the shell commands are working and producing the expected output files
How many jobs?
If we asked Snakemake to run kallisto_quant on all three of the reference samples (ref1, ref2, ref3), how many jobs would that be in total?
10 in total:
- 3 * kallisto_quant
- 6 * trimreads
- 1 * kallisto_index
- 0 * countreads
- 0 * calculate_difference
Snakemake is lazy, and laziness is good
For the last few episodes, we’ve told you to run Snakemake like this:
As a reminder, the -j1 flag tells Snakemake to run one
job at a time, and -p is to print out the shell commands
before running them.
The -F flag turns on forceall mode, and in
normal usage you don’t want this.
At the end of the last chapter, we generated some kallisto results by running:
Now try without the -F option. Assuming that the output
files are already created, you’ll see this:
BASH
$ snakemake -j1 -p kallisto.temp33_1/abundance.h5
Building DAG of jobs...
Nothing to be done.
Complete log: /home/zenmaster/data/yeast/.snakemake/log/2021-04-23T172441.519500.snakemake.logIn normal operation, Snakemake only runs a job if:
- A target file you explicitly requested to make is missing
- An intermediate file is missing and it is needed in the process of making a target file
- Snakemake can see an input file which is newer than an output file
- A rule definition or configuration has changed since the output file was created
The last of these relies on a ledger that Snakemake saves into the
.snakemake directory.
Let’s demonstrate each of these in turn, by altering some files and
re-running Snakemake without the -F option.
This just re-runs kallisto_quant - the final step.
“Nothing to be done” - some intermediate output is missing but Snakemake already has the file you are telling it to make, so it doesn’t worry.
The touch command is a standard Linux command which sets
the timestamp of the file, so now the transcriptome looks to Snakemake
as if it was just modified.
Snakemake sees that one of the input files used in the process of
producing kallisto.temp33_1/abundance.h5 is newer than the
existing output file, so it needs to run the kallisto index and
kallisto quant steps again. Of course, the kallisto
quant step needs the trimmed reads which we deleted earlier, so now
the trimming step is re-run also.
Explicitly telling Snakemake what to re-run
The default timestamp-based logic is really useful when you want to:
- Change or add some inputs to an existing analysis without re-processing everything
- Continue running a workflow that failed part-way
In most cases you can also rely on Snakemake to detect when you have edited a rule, but sometimes you need to be explicit, for example if you have updated an external script or changed a setting which Snakemake doesn’t see.
The -R flag allows you to explicitly tell Snakemake that
a rule has changed and that all outputs from that rule need to be
re-evaluated.
Note on -R
Due to a quirk of the way Snakemake parses command-line options, you
need to make sure there are options after the -R ...,
before the list of target outputs. If you don’t do this, Snakemake will
think that the target files are instead items to add to the
-R list, and then when building the DAG it will just try to
run the default rule.
The easiest way is to put the -p flag before the target
outputs. Then you can list multiple rules to re-run, and also multiple
targets, and Snakemake can tell which is which.
BASH
$ snakemake -j1 -R trimreads kallisto_index -p kallisto.temp33_1/abundance.h5 kallisto.temp33_2/abundance.h5The reason for using the -p flag specifically is that
you pretty much always want this option.
The -f flag specifies that the target outputs named on
the command line should always be regenerated, so you can use this to
explicitly re-make specific files.
This always re-runs kallisto_quant, regardless of whether
the output file is there already. For all intermediate outputs,
Snakemake applies the default timestamp-based logic. Contrast with
-F which runs the entire DAG every time.
Visualising the DAG
Snakemake can draw a picture of the DAG for you, if you run it like this:
Using the --dag option implicitly activates the
-n (dry-run) option so that Snakemake will not actually run
any jobs, it will just print the DAG and stop. Snakemake prints the DAG
in a text format so we use the gm command to make this into
a picture and show it on the screen.
Note on gm display
The gm command is provided by the GraphicsMagick toolkit. On
systems where gm will not display an image directly, you
may instead save it to a PNG file. You will need the dot
program from the GraphViz package
installed.
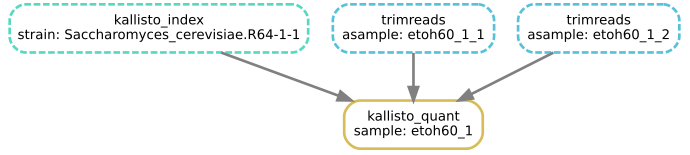
The boxes drawn with dotted lines indicate steps that are not to be run, as the output files are already present and newer than the input files.
Visualising the effect of the -R
and -f flags
Run kallisto_quant on the first of the
etoh60 samples, then use the --dag option
as shown above to check:
How many jobs will run if you ask again to create this output with no
-f,-For-Roptions?How many if you use the
-foption?How many if you use the
-R trimreadsoption?How many if you edit the Snakefile so that the
qual_thresholdfortrimreadsis “22”, rather than “20”?
This is a way to make the Kallisto result in the first place:
- This command should show four boxes, but all are dotted so no jobs are actually to be run.
The
-fflag re-runs only the job to create the output file, so in this case one box is solid, and only that job will run.With
-R trimreads, the two trimreads jobs will re-run, and Snakemake sees that this also requires re-running kallisto_quant, so the answer is 3.
If you see a message like the one below, it’s because you need to put
an option after trimreads or else Snakemake gets confused
about what are parameters of -R, and what things are
targets.
ERROR
WorkflowError:
Target rules may not contain wildcards.- Editing the Snakefile has the same effect as forcing the trimreads rule to re-run, so again there will be three jobs to be run from the DAG.
With older versions of Snakemake this would not be auto-detected, and
in fact you can see this behaviour if you remove the hidden
.snakemake directory. Now Snakemake has no memory of the
rule change so it will not re-run any jobs unless explicitly told
to.
Removing files to trigger reprocessing
In general, getting Snakemake to re-run things by removing files is a
bad idea, because it’s easy to forget about intermediate files that
actually contain stale results and need to be updated. Using the
-R flag is simpler and more reliable. If in doubt, and if
it will not be too time consuming, keep it simple and just use
-F to run the whole workflow from scratch.
For the opposite case where you want to avoid re-running particular
steps, see the ‑‑touch option of Snakemake mentioned later in the course.
- A job in Snakemake is a rule plus wildcard values (determined by working back from the requested output)
- Snakemake plans its work by arranging all the jobs into a DAG (directed acyclic graph)
- If output files already exist, Snakemake can skip parts of the DAG
- Snakemake compares file timestamps and a log of previous runs to determine what need regenerating
