Chaining rules
Last updated on 2025-02-25 | Edit this page
Overview
Questions
- How do I combine rules into a workflow?
- How can I make a rule with multiple input files?
Objectives
- Use Snakemake to filter and then count the sequences in a FASTQ file
- Understand how rules are linked by filename patterns
- Add a rule that calculates the number of reads filtered out
For reference, this is the Snakefile you should have to start the episode.
A pipeline of multiple rules
Our goal at this point is to apply a quality filter to our reads and to see how many reads are discarded by that filter for any given sample. We are not quite there yet, but we do have a countreads rule and a trimreads rule. Following the previous chapter, the contents of the Snakefile should be:
# New generic read counter
rule countreads:
output: "{myfile}.fq.count"
input: "reads/{myfile}.fq"
shell:
"echo $(( $(wc -l <{input}) / 4 )) > {output}"
# Trim any FASTQ reads for base quality
rule trimreads:
output: "trimmed/{myfile}.fq"
input: "reads/{myfile}.fq"
shell:
"fastq_quality_trimmer -t 20 -l 100 -o {output} <{input}"The missing piece is that there is no way to count the reads in the trimmed file. The countreads rule only takes input reads from the reads directory, whereas the trimreads rule puts all results into the trimmed directory.
To fix this, we could move the trimmed reads into the reads directory, or add a second read-counting rule, but the most elegant solution here is to make the countreads rule even more generic, so it can count everything.
# New even-more-generic read counter
rule countreads:
output: "{indir}.{myfile}.fq.count"
input: "{indir}/{myfile}.fq"
shell:
"echo $(( $(wc -l <{input}) / 4 )) > {output}"Now, the rule no longer requires the input files to be in the “reads”
directory. The directory name has been replaced by the
{indir} wildcard. We can request Snakemake to create a file
following this new output pattern:
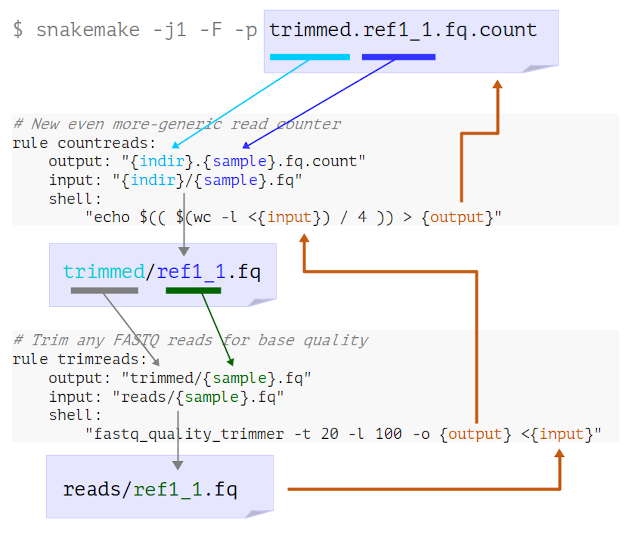
Look at the logging messages that Snakemake prints in the terminal. What has happened here?
- Snakemake looks for a rule to make
trimmed.ref1_1.fq.count - It determines that “countreads” can make this if
indir=trimmedandmyfile=ref1_1 - It sees that the input needed is therefore
trimmed/ref1_1.fq - Snakemake looks for a rule to make
trimmed/ref1_1.fq - It determines that “trimreads” can make this if
myfile=ref1_1 - It sees that the input needed is therefore
reads/ref1_1.fq - Now Snakemake has reached an available input file, it runs both steps to get the final output
Here’s a visual representation of this process:
This, in a nutshell, is how we build workflows in Snakemake.
- Define rules for all the processing steps
- Choose
inputandoutputnaming patterns that allow Snakemake to link the rules - Tell Snakemake to generate the final output files
If you are used to writing regular scripts this takes a little getting used to. Rather than listing steps in order of execution, you are always working backwards from the final desired result. The order of operations is determined by applying the pattern matching rules to the filenames, not by the order of the rules in the Snakefile.
Choosing file name patterns
Chaining rules in Snakemake is a matter of choosing filename patterns that connect the rules. There’s something of an art to it, and most times there are several options that will work, but in all cases the file names you choose will need to be consistent and unabiguous.
Seeing how many reads were discarded
How many reads were removed?
How many reads were removed from the ref1 sample by the filtering step?
After generating both the trimmed and untrimmed .count
files we can get this information.
BASH
$ snakemake -j1 -F -p reads.ref1_1.fq.count trimmed.ref1_1.fq.count
$ head *.ref1_1.fq.count
==> reads.ref1_1.fq.count <==
14677
==> trimmed.ref1_1.fq.count <==
14278Subtracting these numbers shows that 399 reads have been removed.
To finish this part of the workflow we will add a third rule to
perform the calculation for us. This rule will need to take both of the
.count files as inputs. We can use the arithmetic features
of the Bash shell to do the subtraction.
Check that this command runs in your terminal. Take care to get the symbols and spaces all correct. As with the original countreads rule, this shell syntax may well be unfamiliar to you, but armed with this working command we can simply substitute the names of any two files we want to compare.
We can put the shell command into a rule.
rule calculate_difference:
output: "ref1_1.reads_removed.txt"
input:
untrimmed = "reads.ref1_1.fq.count",
trimmed = "trimmed.ref1_1.fq.count",
shell:
"echo $(( $(<{input.untrimmed}) - $(<{input.trimmed}) )) > ref1_1.reads_removed.txt"Note that:
- The above rule has two inputs, trimmed and untrimmed
- We can choose what to call the individual inputs, so use descriptive names
- There is a newline after
input:and the next two lines are indented - The
=and,symbols are needed - You can leave off the final comma, but it’s generally easier to just put one on every line
- We refer to the input file names as
{input.untrimmed}and{input.trimmed} - There is only one output, but we can have multiple named outputs too.
Making this rule generic
Alter the above rule to make it generic by adding suitable wildcards. Use the generic rule to calculate the number of reads removed from the etoh60_1_1.fq input file.
rule calculate_difference:
output: "{myfile}.reads_removed.txt"
input:
untrimmed = "reads.{myfile}.fq.count",
trimmed = "trimmed.{myfile}.fq.count",
shell:
"echo $(( $(<{input.untrimmed}) - $(<{input.trimmed}) )) > {output}"Here, I’ve chosen to use the wildcard name {myfile}
again, but you can use any name you like. We do also need to ensure that
the output file is referenced using the {output}
placeholder.
Processing all the inputs
It would be nice to have Snakemake run this automatically for all our samples. We’ll see how to do this later, in episode 6.
Outputs first?
The Snakemake approach of working backwards from the desired output
to determine the workflow is why we’re putting the output
lines first in all our rules - to remind us that these are what
Snakemake looks at first!
Many users of Snakemake, and indeed the official documentation,
prefer to have the input first, so in practise you should
use whatever order makes sense to you.
For reference, this is a Snakefile incorporating the changes made in this episode.
- Snakemake links up rules by iteratively looking for rules that make missing inputs
- Careful choice of filenames allows this to work
- Rules may have multiple named input files (and output files)
