Introduction to Pangenomics
Overview
Teaching: 10 min
Exercises: 15 minQuestions
What is a pangenome?
What are the components of a pangenome?
Objectives
Gain insights into the origin and significance of pangenomics, and comprehend its fundamental principles.
Acquire a comprehensive understanding of pangenome structure, including the classification of pangenomes based on their element composition.
Unveiling the Genomic Complexity: The Pangenome Paradigm
A brief history of the concept “Pangenome”
The concept of Pangenomics was created by Tettelin et al., whose goal was to develop a vaccine against Group B Streptococcus (GBS, or Streptococcus agalactiae), a human pathogen causing neonatal infections. Previous to this, reverse vaccinology had been successfully applied to Neisseria meningitidis using a single genome. However, in the case of S. agalactiae, two complete sequences were available when the project started. These initial genomic studies revealed significant variability in gene content among closely related S. agalactiae isolates, challenging the assumption that a single genome could represent an entire species. Consequently, the collaborative team decided to sequence six additional genomes, representing the major disease-causing serotypes. The comparison of these genomes confirmed the presence of diverse regions, including differential pathogenicity islands, and revealed that the shared set of genes accounted for only about 80% of an individual genome. The existence of broad genomic diversity prompted the question of how many sequenced genomes are needed to identify all possible genes harbored by S. agalactiae. Motivated by the goal of identifying vaccine candidates, the collaborators engaged in active discussions, scientific drafts, and the development of a mathematical model to determine the optimal number of sequenced strains. And, this is how these pioneering authors introduced the revolutionary concept of the pangenome in 2005.
The term “pangenome” is a fusion of the Greek words pan, which means ‘whole’, and genome, referring to the complete set of genes in an organism. By definition, a pangenome represents the entirety of genes present in a group of organisms, typically a species. Notably, the pangenome concept extends beyond bacteria and can be applied to any taxa of interest, including humans, animals, plants, fungi, archea, or viruses.
Pizza pangenomics
Do you feel confused about what a pangenome is? Look at this analogy!
Solution
Imagine you’re on a mission to open the finest pizza restaurant, aiming to offer your customers a wide variety of pizzas from around the world. To achieve this, you set out to gather all the pizza recipes ever created, including margherita, quattro formaggi, pepperoni, Hawaiian, and more. As you examine these recipes, you begin to notice that certain ingredients appear in multiple pizzas, while others are unique to specific recipes.
In this analogy, the pizza is your species of interest. Your collection of pizza recipes represents the pangenome, which encompasses the entire diversity of pizzas. Each pizza recipe corresponds to one genome, and its ingredients to genes. The “same” ingredient (such as tomato) in the different recipes, would constitute a gene family. Within these common ingredients, there may be variations in brands or preparation styles, reflecting the gene variation within the gene families.
As you continue to add more recipes to your collection, you gain a better understanding of the vast diversity in pizza-making techniques. This enables you to fulfill your objective of offering your customers the most comprehensive and diverse pizza menu.
The components and classification of pangenomes
The pangenome consists of two primary components or partitions: core genome and accessory genome. The core genome comprises gene families that are present in all the genomes being compared, while the accessory genome consists of gene families that are not shared by all genomes. Within the accessory genome, we can further distinguish two partitions, the shell genome, which encompasses the gene families found in the majority of genomes, and the cloud genome, which comprises gene families present in only a minority of genomes. It is worth mentioning that, the specific percentages used to define these partitions may vary across different pangenome analysis software and among researchers. Additional terms such as persistent genome and soft-core genome are also commonly used in the field for the groups of genes that are in almost all of the genomes considered.
Exercise 1(Begginer): Pizza pangenomics
What are the partitions in your pizza pangenome?
Solution
During your expedition of gathering pizza recipes you find that only flour, water, salt, and yeast are in all the recipes, this is your pizza core genome. Since the vast majority of pizzas have tomato sauce and cheese (believe it or not, there are white pizzas without tomato sauce and pizzas without cheese), you put the tomato and the cheese in the soft-core genome. Other ingredients like basil and olive oil are very common so they go to the shell genome, and finally the weirdos like pineapple go to the cloud genome.
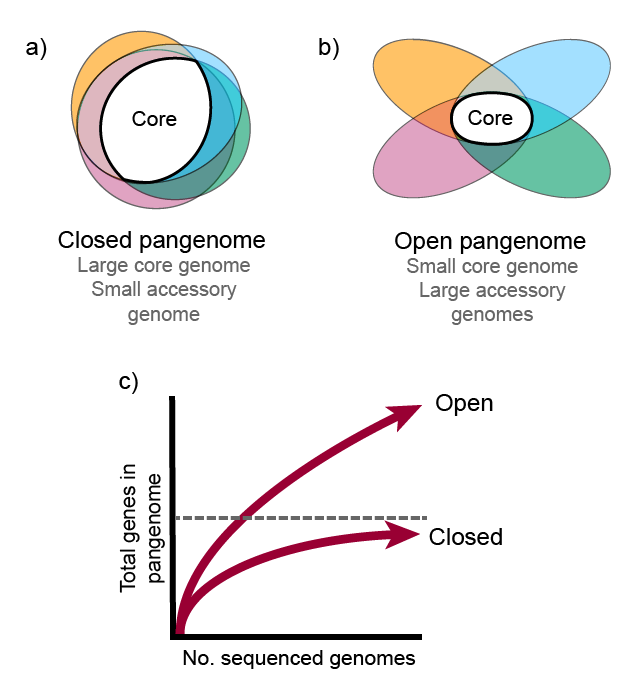
The concept of pangenome encompasses two types: the open pangenome and the closed pangenome. An open pangenome refers to a scenario where the addition of new genomes to a species leads to a significant increase in the total number of gene families. This indicates a high level of genomic diversity and the potential acquisition of new traits with each newly included genome. In contrast, a closed pangenome occurs when the incorporation of new genomes does not contribute significantly to the overall gene family count. In a closed pangenome, the gene family pool remains relatively stable and limited, indicating a lower degree of genomic diversity within the species.
To understand these concepts better, we can visualize the pangenome as a matrix representing the presence (1) or absence (0) of orthologous gene families. The columns represent the gene families, while the rows represent the genomes added to the pangenome. In an open pangenome, the number of columns increases significantly as new genomes are added. Conversely, in a closed pangenome, the number of columns remains relatively unchanged as the number of genomes increases. This suggests that the species maintains a consistent set of gene families. Consequently, the size of the core genome in a closed pangenome closely matches that of a single complete genome of the species. In contrast, the core size of an open pangenome is relatively smaller compared to the size of an individual genome.
In summary, the terms “open pangenome” and “closed pangenome” describe the dynamic nature of gene content in a species, with the former signifying an expanding gene family pool and the latter representing a more stable gene family repertoire.
Exercise 2(Begginer): Open or closed?
The size of a pangenome can be influenced by factors such as the extent of gene transfer, interactions with other species in the environment they co-habit, the diversity of niches inhabited, and the lifestyle of the species, among others.
Considering a human lung pathogen and a soil bacterium, which one do you believe is more likely to have a closed pangenome, characterized by a relatively stable gene pool, and why?
Solution
Based on the assumption that a human lung pathogen may have a more specialized lifestyle and limited exposure to diverse environments, it is likely to possess a closed pangenome. In contrast, a soil bacterium, which encounters a wide range of ecological niches and interacts with various organisms, is more likely to have an open pangenome. However, these assumptions are not always true. Thus, further investigation and analysis are required to confirm these assumptions for the different species of interest.
Know more
If you want to read more on pangenomics go to the book The Pangenome edited by Tettelin and Medini.
Genome database for this workshop
Description of the dataset
In this lesson, we will follow a standard pangenomics pipeline that involves genomic annotation, clustering of genes to identify orthologous sequences and build the gene families, and analyzing the pangenome partitions and openness. To illustrate these concepts, we will work with a dataset consisting of eight strains of Streptococcus agalactiae as included in the pioneering pangenome study by Tettelin et al., 2005 (See the Table below).
We already have the genomes of strains 18RS21 and H36B available in our pan_workshop/data directory. However, the remaining strains will be downloaded and annotated in the upcoming episodes, allowing us to explore the complete dataset.
General description of the S. agalactiae genomes
| Strain | Host | Serotype |
|---|---|---|
| S. agalactiae 18RS21 | Human | II |
| S. agalactiae 515 | Human | Ia |
| S. agalactiae A909 | Human | Ia |
| S. agalactiae CJB111 | Human | V |
| S. agalactiae COH1 | Human | III |
| S. agalactiae H36B | Human | Ib |
| S. agalactiae NEM316 | Human | III |
| S. agalactiae 2603V/R | Human | V |
Prepare your genome database
Make sure you have the
pan_workshop/directory in your home directory. If you do not have it, you can download it with the following instructions.$ cd ~ #Make sure you are in the home directory $ wget https://zenodo.org/record/7974915/files/pan_workshop.zip?download=1 #Download the `zip` file. $ unzip 'pan_workshop.zip?download=1' $ rm 'pan_workshop.zip?download=1'
Key Points
A pangenome encompasses the complete collection of genes found in all genomes within a specific group, typically a species.
Comparing the complete genome sequences of all members within a clade allows for the construction of a pangenome.
The pangenome consists of two main components: the core genome and the accessory genome.
The accessory genome can be further divided into the shell genome and the cloud genome.
In an open pangenome, the size of the pangenome significantly increases with the addition of each new genome.
In a closed pangenome, only a few gene families are added to the pangenome when a new genome is introduced.