Linear regression with one continuous and one categorical explanatory variable
Overview
Teaching: 15 min
Exercises: 20 minQuestions
How can we visualise the relationship between three variables, two of which are continuous and one of which is categorical, in R?
How can we fit a linear regression model to this type of data in R?
How can we obtain and interpret the model parameters?
How can we visualise the linear regression model in R?
Objectives
Explore the relationship between a continuous dependent variable and two explanatory variables, one continuous and one categorical, using ggplot2.
Fit a linear regression model with one continuous explanatory variable and one categorical explanatory variable using lm().
Use the jtools package to interpret the model output.
Use the interactions package to visualise the model output.
In this episode we will learn to fit a linear regression model when we have two explanatory variables: one continuous and one categorical. Before we fit the model, we can explore the relationship between our variables graphically. We are asking the question: does the relationship between the continuous explanatory variable and the response variable differ between the levels of the categorical variable?
Exploratory plots
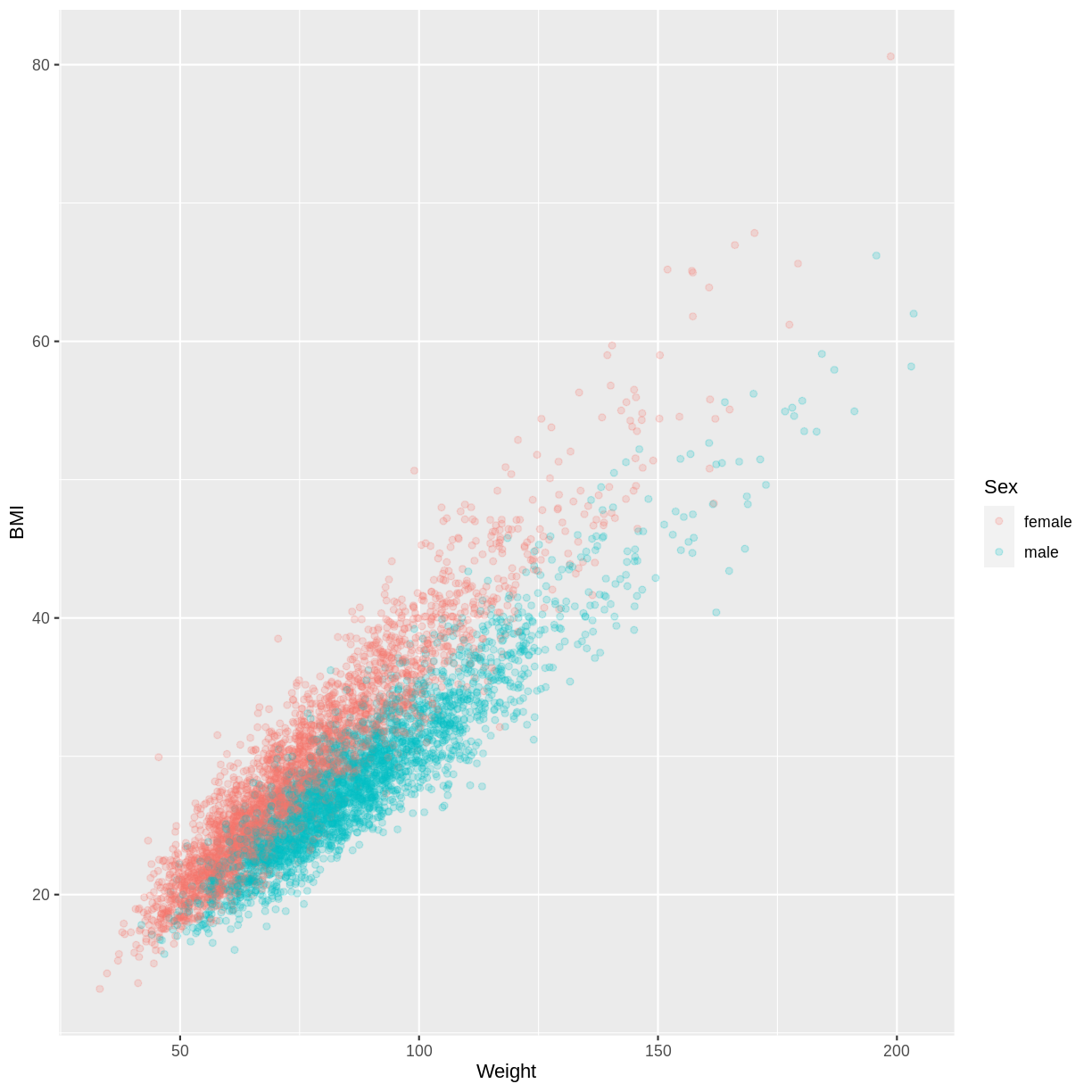
Let us take the response variable BMI, the continuous explanatory variable Weight and the categorical explanatory variable Sex as an example. The code below subsets our data for individuals who are older than 17 years with filter(). The plotting is then initiated using ggplot(). Inside aes(), we select the response variable with y = BMI, the continuous explanatory variable with x = Weight and the categorical explanatory variable with colour = Sex. As a consequence of the final argument, the points produced by geom_point() are coloured by Sex. Finally, we include opacity using alpha = 0.2, which allows us to distinguish areas dense in points from sparser areas. Note that RStudio returns the warning “Removed 320 rows containing missing values (geom_point).” - this reflects that 320 participants had missing BMI and/or Weight data.
The plot suggests that on average, participants classed as female have a higher BMI than participants classed as male, for any particular value of Weight. We might therefore want to account for this in our model by having separate intercepts for the levels of Sex.
dat %>%
filter(Age > 17) %>%
ggplot(aes(x = Weight, y = BMI, colour = Sex)) +
geom_point(alpha = 0.2)
Exercise
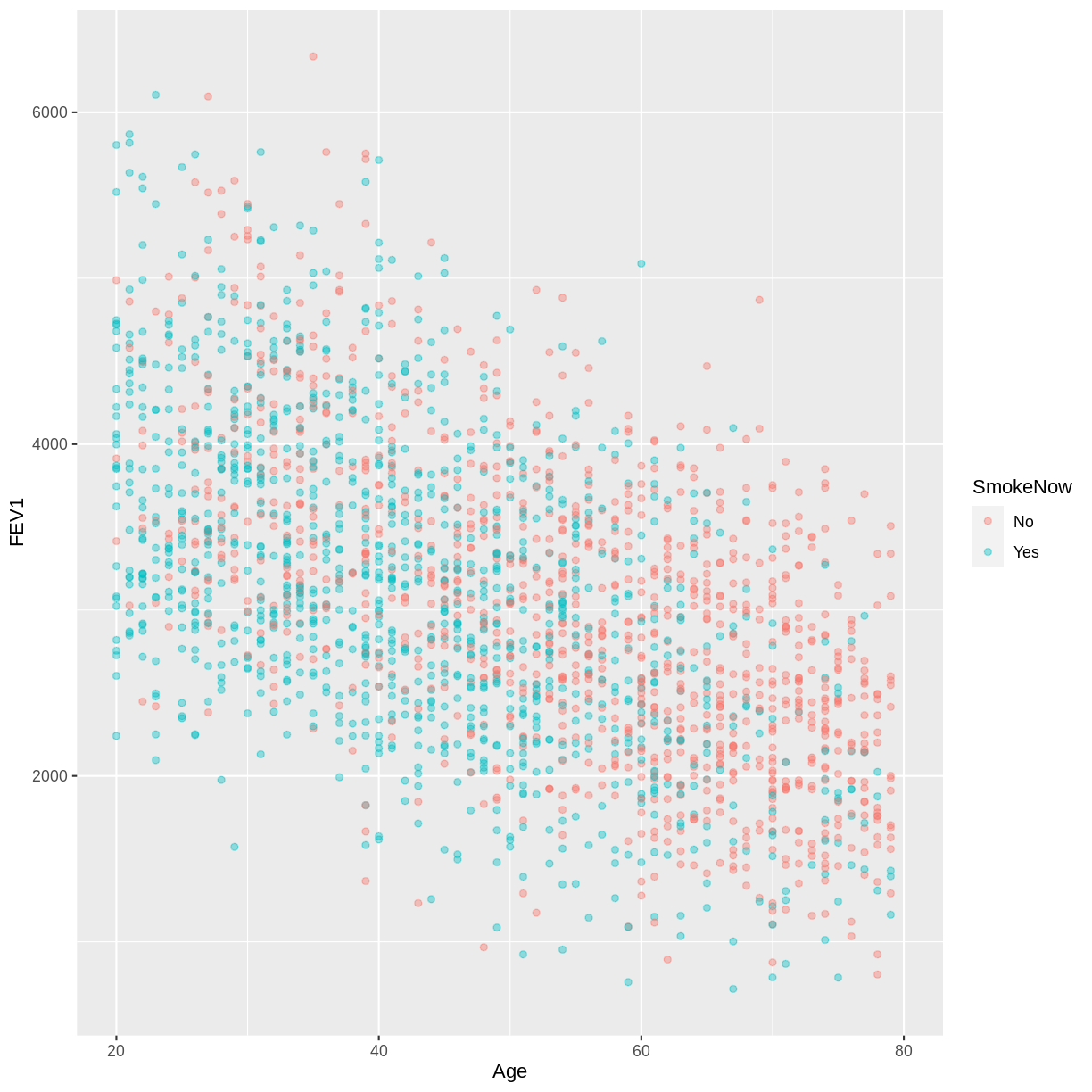
You have been asked to model the relationship between
FEV1andAgein the NHANES data, splitting observations by whether participants are active or ex smokers (SmokeNow). Use theggplot2package to create an exploratory plot, ensuring that:
- NAs are discarded from the
SmokeNowvariable.- FEV1 (
FEV1) is on the y-axis and Age (Age) is on the x-axis.- These data are shown as a scatterplot, with points coloured by
SmokeNowand an opacity of 0.4.Solution
dat %>% drop_na(SmokeNow) %>% ggplot(aes(x = Age, y = FEV1, colour = SmokeNow)) + geom_point(alpha = 0.4)
Fitting and interpreting a multiple linear regression model
We fit the model using lm(). With formula = BMI ~ Weight + Sex we specify that our model has two explanatory variables, Weight and Sex.
BMI_Weight_Sex <- dat %>%
filter(Age > 17) %>%
lm(formula = BMI ~ Weight + Sex)
The linear model equation associated with this model has the general form
[E(y) = \beta_0 + \beta_1 \times x_1 + \beta_2 \times x_2.]
Notice that we now have the additional parameters, $\beta_2$ and $x_2$, in comparison to the simple linear regression model equation. In the context of our model, $\beta_1$ is the parameter for the effect of Weight and $\beta_2$ is the parameter for the effect of Sex. Recall from the simple linear regression lesson that a categorical variable has a baseline level in R. The parameter associated with the categorical variable then estimates the difference in the outcome variable in a group different from the baseline. Since “f” precedes “m” in the alphabet, R takes female as the baseline level. Therefore, $\beta_2$ estimates the difference in BMI between males and females, given a particular value for Weight. $\beta_2$ is only included when $x_2 = 1$, i.e. when the Sex of a participant is male. This results in separate intercepts for the levels of Sex: the intercept for females is given by $\beta_0$ and the intercept for males is given by $\beta_0 + \beta_2$. In this model, the effect of Weight is given by $\beta_1$, irrespective of the Sex of a participant. This results in equal slopes across the levels of Sex.
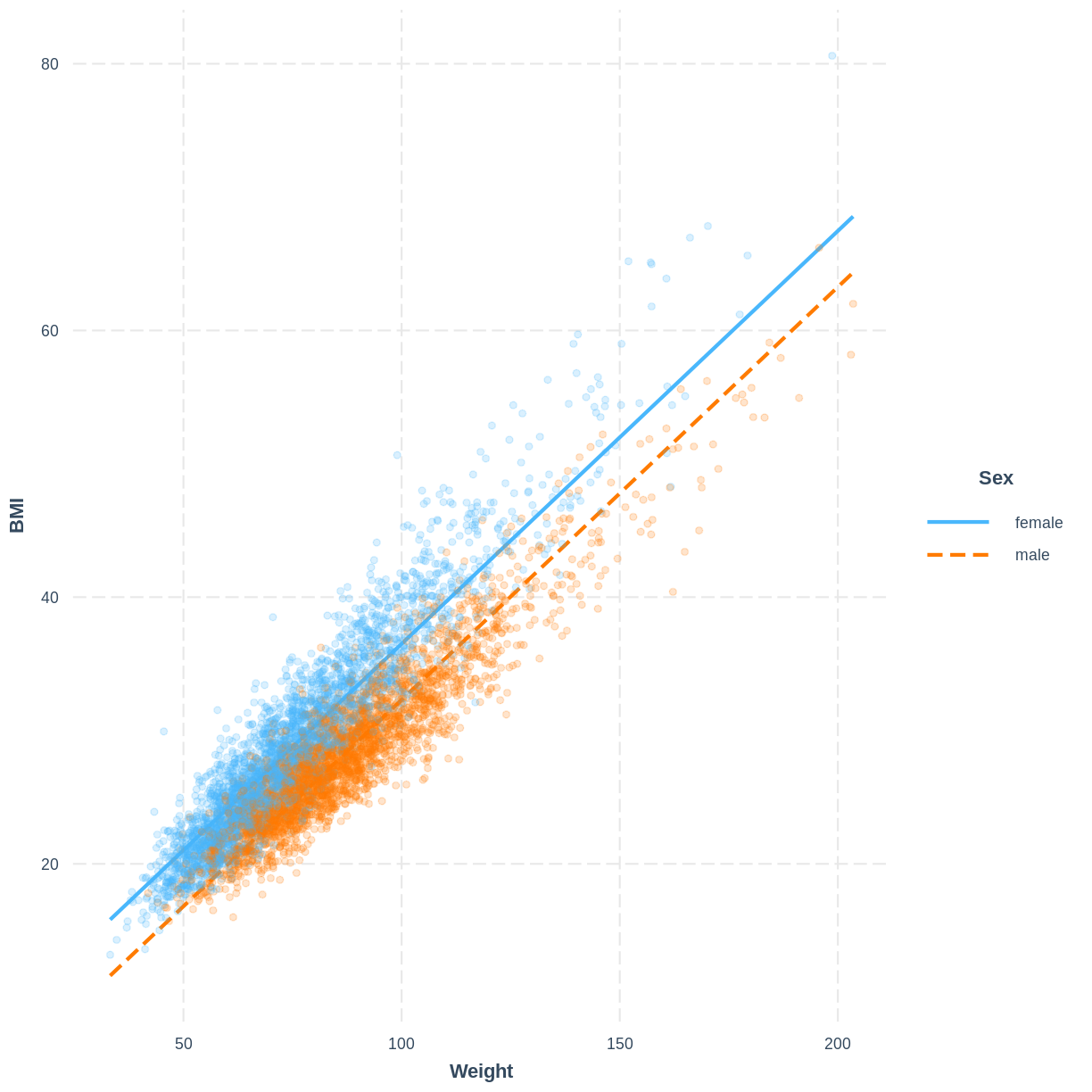
The model parameters can be obtained using summ() from the jtools package. We include 95% confidence intervals for the parameter estimates using confint = TRUE and three digits after the decimal point using digits = 3. The intercept when Sex = female equals $5.538$ and the intercept when Sex = male equals $5.538 - 4.202 = 1.336$. Notice that in this model, the effect of Weight is $0.310$, regardless of Sex. The model equation can therefore be updated to be:
[E(\text{BMI}) = 5.538 + 0.310 \times \text{Weight} -4.202 \times x_2,]
where $x_2 = 1$ for Sex = male and $0$ otherwise.
summ(BMI_Weight_Sex, confint = TRUE, digits = 3)
MODEL INFO:
Observations: 6177 (320 missing obs. deleted)
Dependent Variable: BMI
Type: OLS linear regression
MODEL FIT:
F(2,6174) = 19431.981, p = 0.000
R² = 0.863
Adj. R² = 0.863
Standard errors: OLS
--------------------------------------------------------------
Est. 2.5% 97.5% t val. p
----------------- -------- -------- -------- --------- -------
(Intercept) 5.538 5.289 5.787 43.656 0.000
Weight 0.310 0.307 0.313 196.965 0.000
Sexmale -4.202 -4.333 -4.071 -63.075 0.000
--------------------------------------------------------------
Exercise
- Using the
lm()command, fit a multiple linear regression of FEV1 (FEV1) as a function of Age (Age), grouped bySmokeNow. Name thislmobjectFEV1_Age_SmokeNow.- Using the
summfunction from thejtoolspackage, answer the following questions:A) What
FEV1does the model predict, on average, for an individual, belonging to the baseline level ofSmokeNow, with anAgeof 0?
B) What is R taking as the baseline level ofSmokeNowand why?
C) By how much isFEV1expected to differ, on average, between the baseline and the alternative level ofSmokeNow?
D) By how much isFEV1expected to differ, on average, for a one-unit difference inAge?
E) Given these values and the names of the response and explanatory variables, how can the general equation $E(y) = \beta_0 + {\beta}_1 \times x_1 + {\beta}_2 \times x_2$ be adapted to represent the model?Solution
FEV1_Age_SmokeNow <- dat %>% lm(formula = FEV1 ~ Age + SmokeNow) summ(FEV1_Age_SmokeNow, confint = TRUE, digits=3)MODEL INFO: Observations: 2265 (7735 missing obs. deleted) Dependent Variable: FEV1 Type: OLS linear regression MODEL FIT: F(2,2262) = 560.807, p = 0.000 R² = 0.331 Adj. R² = 0.331 Standard errors: OLS -------------------------------------------------------------------- Est. 2.5% 97.5% t val. p ----------------- ---------- ---------- ---------- --------- ------- (Intercept) 4928.155 4805.493 5050.816 78.787 0.000 Age -35.686 -37.799 -33.573 -33.117 0.000 SmokeNowYes -249.305 -317.045 -181.564 -7.217 0.000 --------------------------------------------------------------------A) 4928.155 mL
B)No, because “n” precedes “y” in the alphabet.
C) On average, individuals from the two categories are expected to differ by 249.305 mL.
D) It is expected to decrease by 35.686 mL.
E) $E(\text{FEV1}) = 4928.155 - 35.686 \times \text{Age} - 249.305 \times x_2$,
where $x_2 = 1$ for current smokers and $0$ otherwise.
Visualising a multiple linear regression model
The model can be visualised using two lines, representing the levels of Sex. Rather than using effect_plot() from the jtools package, we use interact_plot from the interactions package. We specify the model by its name, the continuous explanatory variable using pred = Weight and the categorical explanatory variable using modx = Sex. We include the data points using plot.points = TRUE and introduce opacity into the data points using point.alpha = 0.2.
Note that R returns the warning “Weight and Sex are not included in an interaction with one another in the model.” - we will learn what interactions are and when they might be appropriate in the next episode. In this scenario we can safely ignore the warning.
interact_plot(BMI_Weight_Sex, pred = Weight, modx = Sex,
plot.points = TRUE, point.alpha = 0.2)
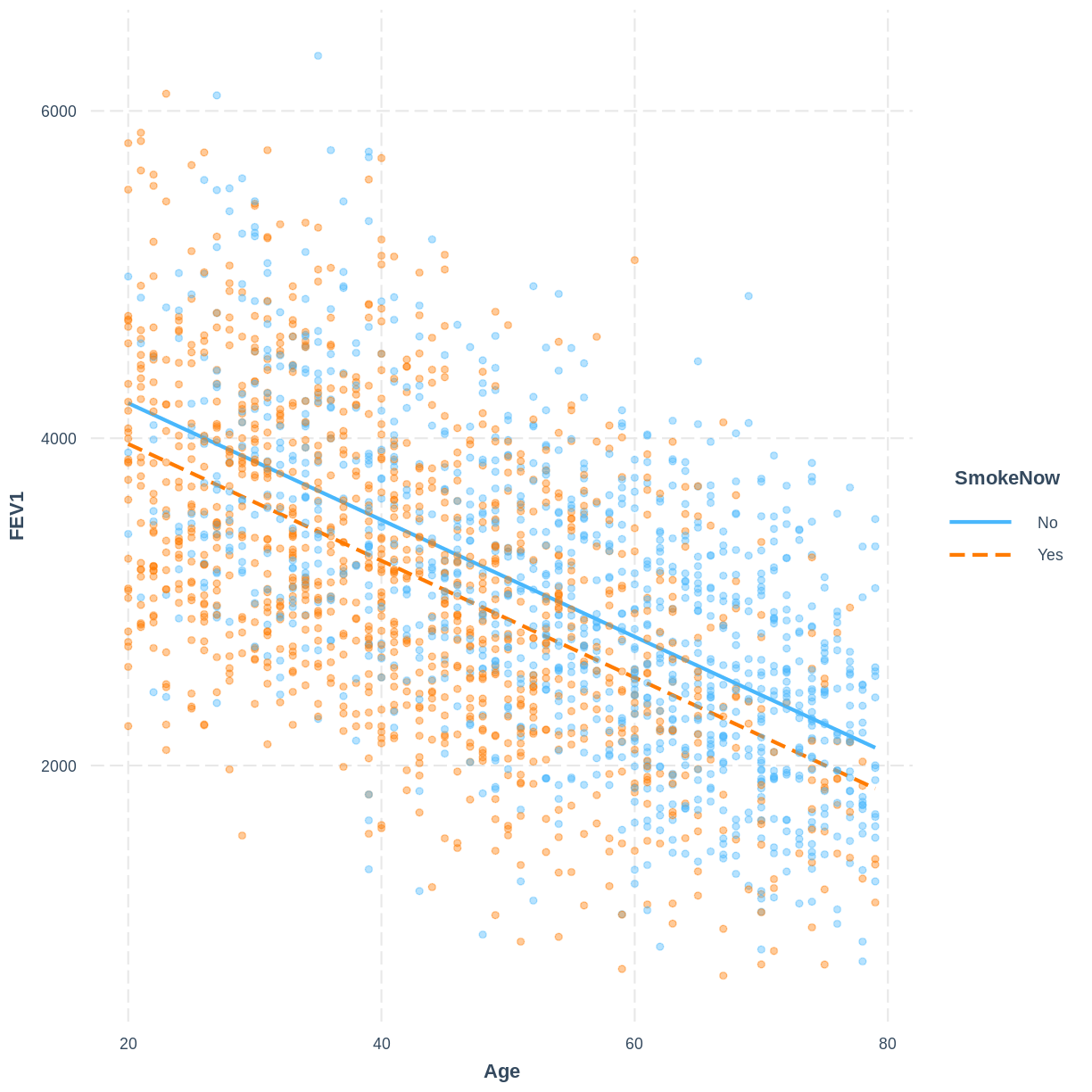
Exercise
To help others interpret the
FEV1_Age_SmokeNowmodel, produce a figure. Make this figure using theinteractionspackage.Solution
interact_plot(FEV1_Age_SmokeNow, pred = Age, modx = SmokeNow, plot.points = TRUE, point.alpha = 0.4)
Key Points
A scatterplot, with points coloured by the levels of a categorical variable, can be used to explore the relationship between two continuous variables and a categorical variable.
The categorical variable can be added to the
formulainlm()using a+.The model output shows separate intercepts for the levels of the categorical variable. The slope across the levels of the categorical variable is held constant.
Parallel lines can be added to the exploratory scatterplot to visualise the linear regression model.
Linear regression including an interaction between one continuous and one categorical explanatory variable
Overview
Teaching: 20 min
Exercises: 30 minQuestions
When is it appropriate to add an interaction to a multiple linear regression model?
How do we add an interaction term in the
lm()command?How do the coefficient estimates given by
summ()relate to the multiple linear regression model equation?How can we visualise the final model in R?
Objectives
Recognise from an exploratory plot when an interaction between a continuous and a categorical explanatory variable is appropriate.
Fit a linear regression model including an interaction between one continuous and one categorical explanatory variable using
lm().Use the jtools package to interpret the model output.
Use the interactions package to visualise the model output.
In the previous episode, we modeled the effect of the continuous explanatory variable as equal across the groups of the categorical explanatory variable. For example, the effect of Weight on BMI was estimated as $0.31$ for both females and males. In this episode we will expand our modelling approach to include an interaction between the continuous and categorical explanatory variables. As a result, not only the intercept but also the coefficient of the explanatory variable will differ across the levels of the categorical variable in our model. This is appropriate when the slope in our scatterplot differs between the levels of the categorical variable.
Visually exploring the need for an interaction
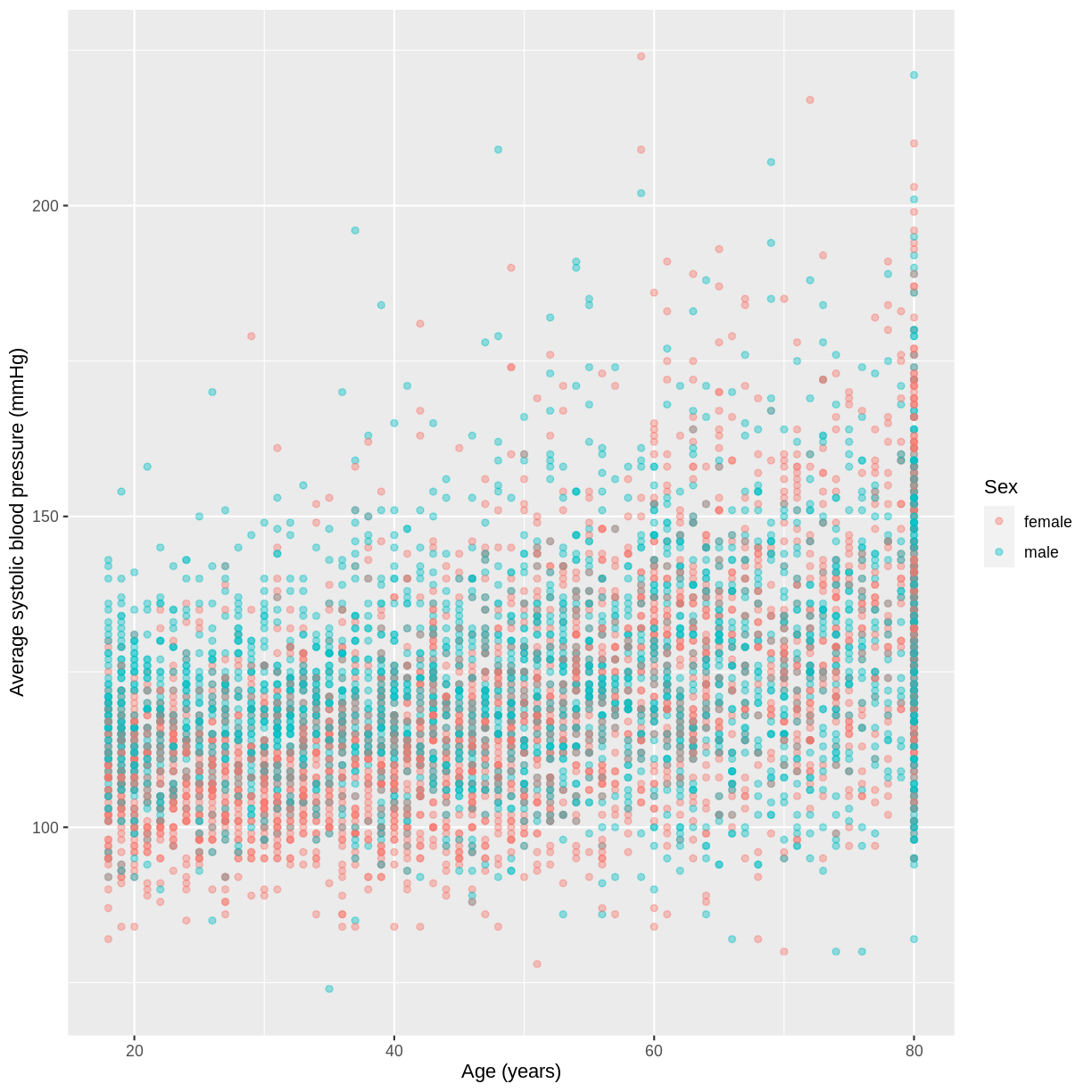
We will use the variables BPSysAve, Age and Sex as an example. Looking at the scatterplot below, for which the data has been filtered to include participants over the age of 17, it seems that the slope for females is greater than for males.
dat %>%
filter(Age > 17) %>%
ggplot(aes(x = Age, y = BPSysAve, colour = Sex)) +
geom_point(alpha = 0.4) +
ylab("Average systolic blood pressure (mmHg)") +
xlab("Age (years)")
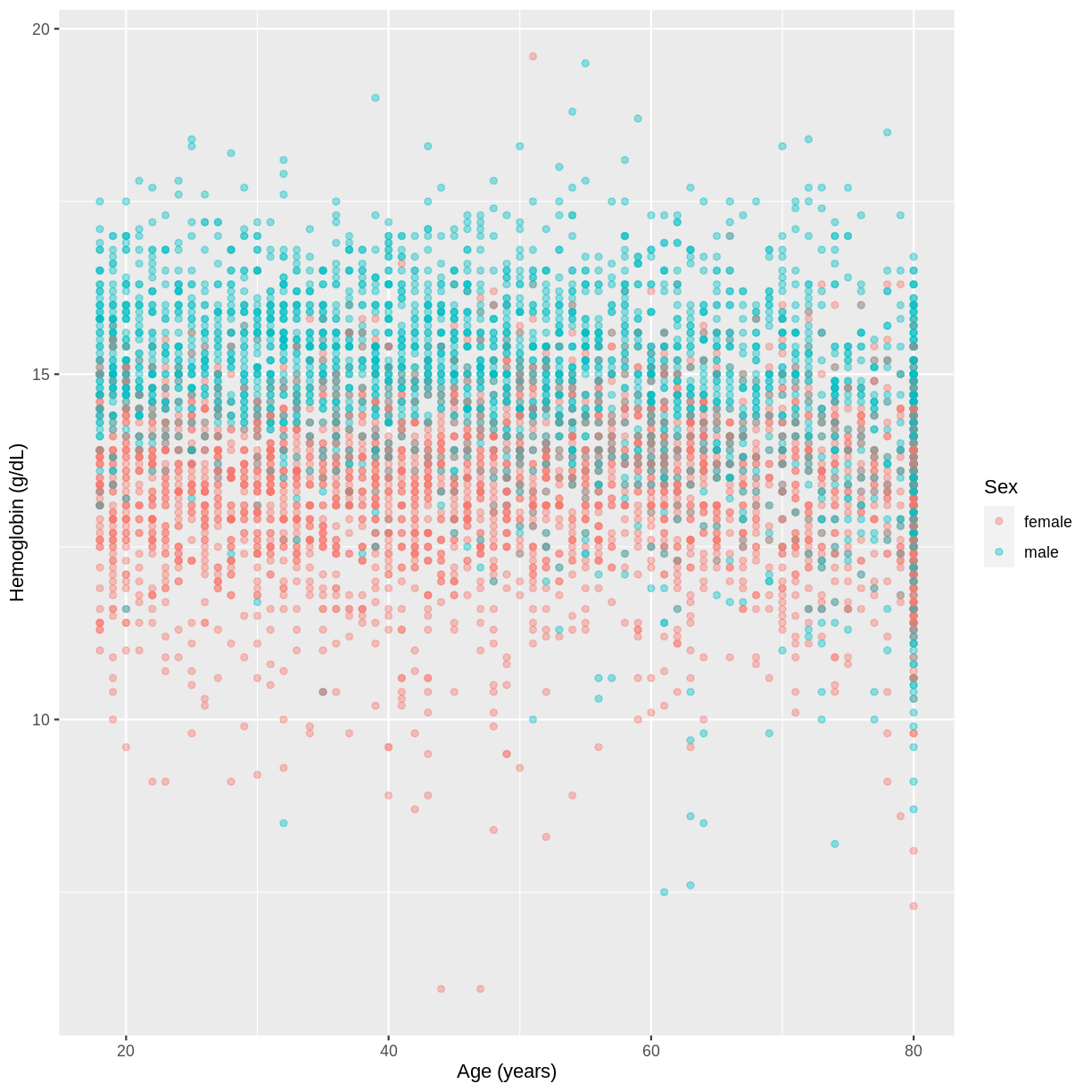
Exercise
You have been asked to model the relationship between
HemoglobinandAgein the NHANES data, splitting observations bySex. Use theggplot2package to create an exploratory plot, ensuring that:
- Only participants aged 18 or over are included.
- Hemoglobin (
Hemoglobin) is on the y-axis and Age (Age) is on the x-axis.- These data are shown as a scatterplot, with points coloured by
Sexand an opacity of 0.4.- The axes are labelled as “Hemoglobin (g/dL)” and “Age (years)”.
Solution
dat %>% filter(Age > 17) %>% ggplot(aes(x = Age, y = Hemoglobin, colour = Sex)) + geom_point(alpha = 0.4) + ylab("Hemoglobin (g/dL)") + xlab("Age (years)")
Fitting and interpreting a multiple linear regression model with an interaction
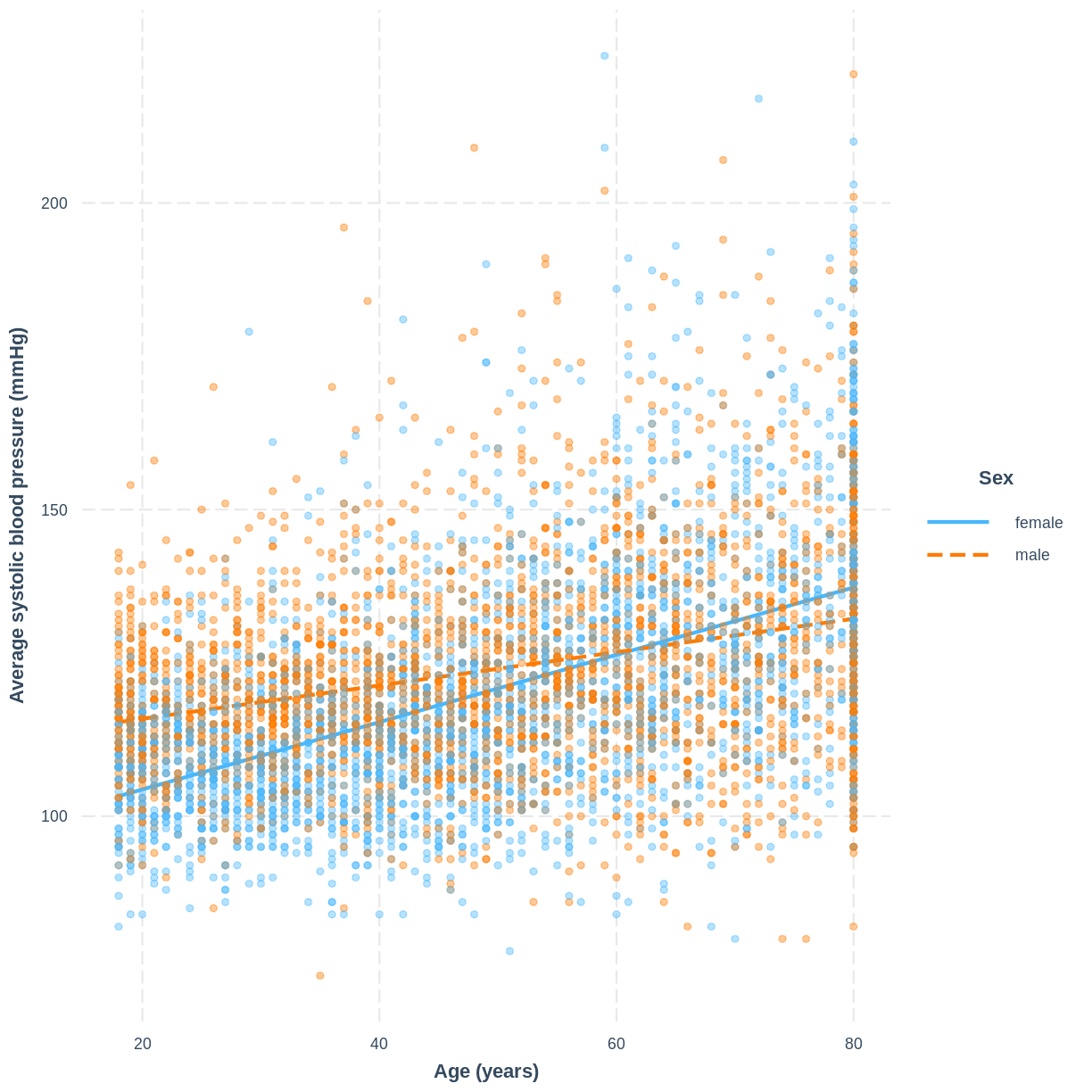
The code for fitting our model is similar to the previous lm() commands. Notice however that instead of a + between the explanatory variables, we use a *. This is how we tell lm() that we want an interaction between the explanatory variables to be included in the model. The interaction allows the effect of Age to differ across the levels of Sex.
In the output from summ() we see two coefficients that relate to the baseline level of Sex: an intercept and the effect for Age. For the alternative level of Sex, we see two further coefficients: the difference in the intercept (Sexmale) and the difference in the slope (Age:Sexmale). The equation for this model therefore becomes:
\(E(\text{Average systolic blood pressure}) = 93.39 + \left(0.55 \times \text{Age}\right) + \left(17.06 \times x_2\right) - \left(0.28 \times x_2 \times \text{Age}\right)\),
where $x_2 = 1$ for male participants and $0$ otherwise.
Since the difference in the intercepts is positive, we expect a greater intercept for males than for females. Furthermore, since the difference in the slopes is negative, we expect a smaller slope for males than for females.
BPSysAve_Age_Sex <- dat %>%
filter(Age > 17) %>%
lm(formula = BPSysAve ~ Age * Sex)
summ(BPSysAve_Age_Sex)
MODEL INFO:
Observations: 5996 (501 missing obs. deleted)
Dependent Variable: BPSysAve
Type: OLS linear regression
MODEL FIT:
F(3,5992) = 552.67, p = 0.00
R² = 0.22
Adj. R² = 0.22
Standard errors: OLS
------------------------------------------------
Est. S.E. t val. p
----------------- ------- ------ -------- ------
(Intercept) 93.39 0.80 116.27 0.00
Age 0.55 0.02 35.63 0.00
Sexmale 17.06 1.15 14.89 0.00
Age:Sexmale -0.28 0.02 -12.68 0.00
------------------------------------------------
Exercise
- Using the
lm()command, fit a multiple linear regression model of Hemoglobin (Hemoglobin) as a function of Age (Age), grouped bySex, including an interaction betweenAgeandSex. Make sure to only include participants who were 18 years or older. Name thelmobjectHemoglobin_Age_Sex.- Using the
summfunction from thejtoolspackage, answer the following questions:A) What concentration of
Hemoglobindoes the model predict, on average, for an individual, belonging to the baseline level ofSex, with anAgeof 0?
B) By how much isHemoglobinexpected to differ, on average, for the alternative level ofSex, at anAgeof 0?
C) By how much isHemoglobinexpected to differ, on average, for a one-unit difference inAgein the baseline level ofSex?
D) By how much isHemoglobinexpected to differ, on average, for a one-unit difference inAgein the alternative level ofSex?
E) Given these values and the names of the response and explanatory variables, how can the general equation $E(y) = \beta_0 + {\beta}_1 \times x_1 + {\beta}_2 \times x_2 + {\beta}_3 \times x_1 \times x_2$ be adapted to represent the model?Solution
Hemoglobin_Age_Sex <- dat %>% filter(Age > 17) %>% lm(formula = Hemoglobin ~ Age * Sex) summ(Hemoglobin_Age_Sex)MODEL INFO: Observations: 5995 (502 missing obs. deleted) Dependent Variable: Hemoglobin Type: OLS linear regression MODEL FIT: F(3,5991) = 1026.31, p = 0.00 R² = 0.34 Adj. R² = 0.34 Standard errors: OLS ------------------------------------------------ Est. S.E. t val. p ----------------- ------- ------ -------- ------ (Intercept) 13.29 0.06 219.70 0.00 Age 0.00 0.00 0.68 0.49 Sexmale 2.76 0.09 31.85 0.00 Age:Sexmale -0.02 0.00 -13.94 0.00 ------------------------------------------------A) 13.29 g/dL.
B) On average, individuals from the two categories are expected to differ by 2.76 g/dL at anAgeof 0.
C) On average, female participants with a one-unit difference inAgeare expected to differ by 0.00 in theirHemoglobinconcentration. So, not at all.
D) On average, male participants with a one-unit difference inAgeare expected to differ by 0.02 in theirHemoglobinconcentration.
E) $E(\text{Hemoglobin}) = 13.29 + 0.00 \times \text{Age} + 2.76 \times x_2 - 0.02 \times \text{Age} \times x_2$, where $x_2 = 1$ for participants of the maleSexand $0$ otherwise.
Visualising a multiple linear regression model with an interaction
We can visualise the model, as before, using the interact_plot() function.
interact_plot(BPSysAve_Age_Sex, pred = Age, modx = Sex,
plot.points = TRUE, point.alpha = 0.4) +
ylab("Average systolic blood pressure (mmHg)") +
xlab("Age (years)")
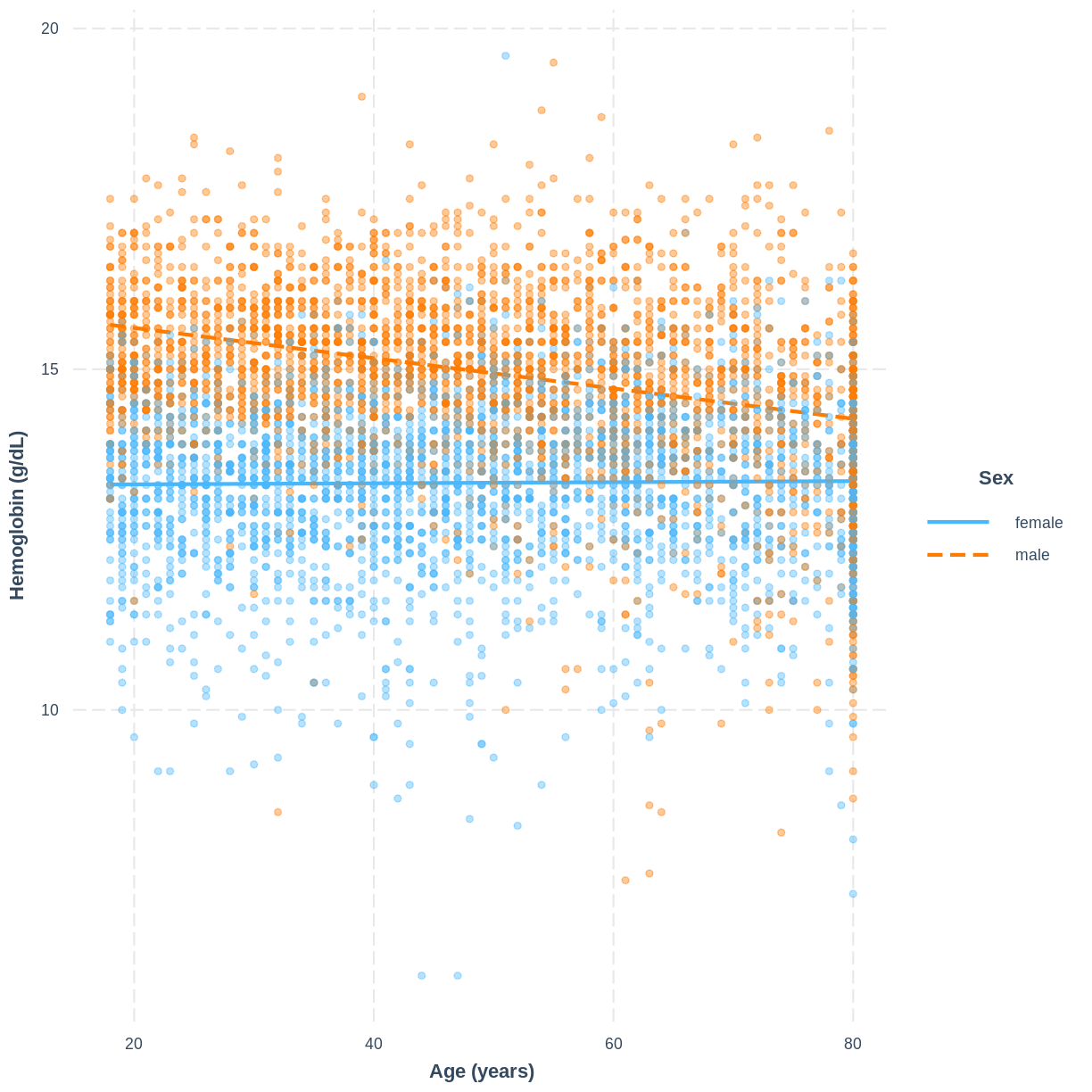
Exercise
To help others interpret the
Hemoglobin_Age_Sexmodel, produce a figure. Make this figure using theinteractionspackage.Solution
interact_plot(Hemoglobin_Age_Sex, pred = Age, modx = Sex, plot.points = TRUE, point.alpha = 0.4) + ylab("Hemoglobin (g/dL)") + xlab("Age (years)")
Key Points
It may be appropriate to include an interaction when the slopes appear to differ across levels of a categorical variable.
Replace
+by*in thelm()command to add an interaction.When an interaction is included, two coefficients relate to differences between the two levels of a categorical variable - one relates to a difference in the intercept, the other to a difference in the slope.
The function
interact_plot()can be used to visualise the model.
Making predictions from a multiple linear regression model
Overview
Teaching: 8 min
Exercises: 12 minQuestions
How can we make predictions using the model equation of a multiple linear regression model?
How can we use R to obtain predictions from a multiple linear regression model?
Objectives
Calculate a prediction from a simple linear regression model using parameter estimates given by the model output.
Use the predict function to generate predictions from a multiple linear regression model.
As with the simple linear regression model, the multiple linear regression model allows us to make predictions. First we will calculate predictions using the model equation. Then we will see how R can calculate predictions for us using the make_predictions() function.
Calculating predictions manually
Let us use the BPSysAve_Age_Sex model from the previous episode. The equation for this model was:
\(E(\text{BPSysAve}) = 93.39 + 0.55 \times \text{Age} + 17.06 \times x_2 - 0.28 \times x_2 \times \text{Age}\),
where $x_2 = 1$ for male participants and $0$ otherwise.
For a 30-year old female, the model predicts an average systolic blood pressure of
\(E(\text{BPSysAve}) = 93.39 + 0.55 \times 30 + 17.06 \times 0 - 0.28 \times 0 \times 30 = 93.39 + 16.5 = 109.89\).
For a 30-year old male, the model predicts an average systolic blood pressure of
\(E(\text{BPSysAve}) = 93.39 + 0.55 \times 30 + 17.06 \times 1 - 0.28 \times 1 \times 30 = 93.39 + 16.5 + 17.06 - 8.4 = 118.55\).
Exercise
Given the
summoutput from ourHemoglobin_Age_Sexmodel, the model can be described as \(E(\text{Hemoglobin}) = 13.29 + 0.00 \times \text{Age} + 2.76 \times x_2 - 0.02 \times x_2 \times \text{Age}\), where $x_2 = 1$ for participants of the maleSexand $0$ otherwise.
- What level of Hemoglobin does the model predict, on average, for an individual of the female
Sexwith anAgeof 40?- What
Hemoglobinis predicted for a male of the sameAge?Solution
For 40-year old females: $13.29 + 0.00 \times 40 + 2.76 \times 0 - 0.02 \times 0 \times 40 = 13.29$.
For 40-year old males: $13.29 + 0.00 \times 40 + 2.76 \times 1 - 0.02 \times 1 \times 40 = 13.29 + 2.76 - 0.8 = 15.25$.
Making predictions using make_predictions()
Using the make_predictions() function brings two advantages. First, when calculating multiple predictions, we are saved the effort of inserting multiple values into our model manually and doing the calculations. Secondly, make_predictions() returns 95% confidence intervals around the predictions, giving us a sense of the uncertainty around the predictions.
To use make_predictions(), we need to create a tibble with the explanatory variable values for which we wish to have mean predictions from the model. We do this using the tibble() function. Note that the column names must correspond to the names of the explanatory variables in the model, i.e. Age and Sex. In the code below, we create a tibble with Age having the values 30, 40, 50 and 60 for females and males. We then provide make_predictions() with this tibble, alongside the model from which we wish to have predictions. By default, make_predicitons() returns
95% confidence intervals for the mean predictions.
From the output we can see that the model predicts an average systolic blood pressure of 109.8596 for a 30-year old female. The confidence interval around this prediction is [109.0593, 110.6599].
BPSysAve_Age_Sex <- dat %>%
filter(Age > 17) %>%
lm(formula = BPSysAve ~ Age * Sex)
predictionDat <- tibble(Age = c(30, 40, 50, 60,
30, 40, 50, 60),
Sex = c("female", "female", "female", "female",
"male", "male", "male", "male"))
make_predictions(BPSysAve_Age_Sex, new_data = predictionDat)
# A tibble: 8 × 5
Age Sex BPSysAve ymin ymax
<dbl> <chr> <dbl> <dbl> <dbl>
1 30 female 110. 109. 111.
2 40 female 115. 115. 116.
3 50 female 121. 120. 121.
4 60 female 126. 126. 127.
5 30 male 119. 118. 119.
6 40 male 121. 121. 122.
7 50 male 124. 123. 125.
8 60 male 127. 126. 127.
Exercise
Using the
make_predictions()function, obtain the expected mean Hemoglobin levels predicted by theHemoglobin_Age_Sexmodel for individuals with anAgeof 20, 30, 40 and 50. Obtain confidence intervals for these predictions. How are these confidence intervals interpreted?Solution
Hemoglobin_Age_Sex <- dat %>% filter(Age > 17) %>% lm(formula = Hemoglobin ~ Age * Sex) predictionDat <- tibble(Age = c(20, 30, 40, 50, 20, 30, 40, 50), Sex = c("female", "female", "female", "female", "male", "male", "male", "male")) make_predictions(Hemoglobin_Age_Sex, new_data = predictionDat)# A tibble: 8 × 5 Age Sex Hemoglobin ymin ymax <dbl> <chr> <dbl> <dbl> <dbl> 1 20 female 13.3 13.2 13.4 2 30 female 13.3 13.3 13.4 3 40 female 13.3 13.3 13.4 4 50 female 13.3 13.3 13.4 5 20 male 15.6 15.5 15.7 6 30 male 15.4 15.3 15.4 7 40 male 15.2 15.1 15.2 8 50 male 14.9 14.9 15.0Recall that 95% of the 95% confidence intervals are expected to contain the population mean. Therefore, we can be fairly confident that the true population means lie somewhere between the bounds of the intervals, assuming that our model is good.
Key Points
Predictions of the mean in the outcome variable can be manually calculated using the model’s equation.
Predictions of multiple means in the outcome variable alongside 95% CIs can be obtained using the
make_predictions()function.
Assessing multiple linear regression model fit and assumptions
Overview
Teaching: 15 min
Exercises: 45 minQuestions
Why is the adjusted R squared used, instead of the standard R squared, when working with multiple linear regression?
What are the six assumptions of multiple linear regression and how are they assessed?
Objectives
Use the adjusted R squared value as a measure of model fit.
Assess whether the assumptions of the multiple linear regression model have been violated.
In this episode we will check the fit and assumptions of multiple linear regression models. We will use the extension to $R^2$ for multiple linear regression models, the adjusted $R^2$ ($R^2_{adj}$). We will also learn to assess the six linear regression assumptions in the case of multiple linear regression.
Measuring fit using $R^2_{adj}$
In the simple linear regression episode
on model assumptions we learned that $R^2$ quantifies
the proportion of variation in the outcome variable explained by the
explanatory variable. In the context of multiple linear regression, a
caveat emerges to $R^2$: adding explanatory variables to our model will
always increase $R^2$, even if the explanatory variables are not related to the response variable. Therefore, when interpreting
the output from summ(), look at the $R^2_{adj}$. This is an $R^2$ corrected
for the number of variables in the model. In some cases, the adjusted and
unadjusted metrics will be (near) equal. In other cases, the differences
will be larger.
Exercise
Find the adjusted R-squared value for the
summoutput of ourHemoglobin_Age_Sexmodel from episode 2. What proportion of variation in hemoglobin is explained by age, sex and their interaction in our model? Does our model account for most of the variation inHemoglobin?Solution
Hemoglobin_Age_Sex <- dat %>% filter(Age > 17) %>% lm(formula = Hemoglobin ~ Age * Sex) summ(Hemoglobin_Age_Sex, confint = TRUE)MODEL INFO: Observations: 5995 (502 missing obs. deleted) Dependent Variable: Hemoglobin Type: OLS linear regression MODEL FIT: F(3,5991) = 1026.31, p = 0.00 R² = 0.34 Adj. R² = 0.34 Standard errors: OLS --------------------------------------------------------- Est. 2.5% 97.5% t val. p ----------------- ------- ------- ------- -------- ------ (Intercept) 13.29 13.18 13.41 219.70 0.00 Age 0.00 -0.00 0.00 0.68 0.49 Sexmale 2.76 2.59 2.93 31.85 0.00 Age:Sexmale -0.02 -0.03 -0.02 -13.94 0.00 ---------------------------------------------------------Since $R^2_{adj} = 0.34$, our model accounts for approximately 34% of the variation in
Hemoglobin. Our model explains 34% of the variation inHemoglobin, which a model that always predicts the mean would not.
Assessing the assumptions of the multiple linear regression model
We learned to assess six assumptions in the simple linear regression episode on model assumptions. These assumptions also hold in the multiple linear regression context, although in some cases their assessment is a little more extensive. Below we will practice assessing these assumptions.
Validity
Recall that the validity assumption states that the model is appropriate for the research question. Validity is assessed through three questions:
A) Does the outcome variable reflect the phenomenon of interest?
B) Does the model include all relevant explanatory variables?
C) Does the model generalise to our case of interest?
Exercise
You are asked to use the
FEV1_Age_SmokeNowmodel to answer the following research question:“Does the effect of age on FEV1 differ by smoking status in US adults?”
Using the three points above, assess the validity of this model for the research question.
Solution
A) The research question is on FEV1, which is our outcome variable. Therefore our outcome variable reflects the phenomenon of interest.
B) Our research question asks whether the effect of age differs by smoking status, which can be tested using an interaction betweenAgeandSmokeNow. Since our model does not include this interaction, our model does not include all relevant explanatory variables.
C) Since the NHANES data was collected from individuals in the US, our model should generalise to our case of interest.
Representativeness
Recall that the representativeness assumption states that the sample is representative of the population to which we are generalising our findings. This assumption is assessed in the same way as for the case of the simple linear regression model, so we will not go through another exercise at this point. Note however that when representativesness is violated, this can sometimes be solved by adding the misrepresented variable to the model as an extra explanatory variable.
Linearity and additivity
Recall that this assumption states that our outcome variable has a linear, additive relationship with the explanatory variables.
The linearity component is assessed in the same way as in the simple
linear regression case. For example, recall that the relationship between
combined diastolic blood pressure (BPDiaAve) and age in months (AgeMonths)
was curved (see this episode from the simple linear regression lesson). Adding a squared term to the model helped us to model this
non-linear relationship. We can add a Sex term to fit separate
non-linear curves to this data:
BPDiaAve_AgeMonthsSQ_Sex <- lm(BPDiaAve ~ AgeMonths + I(AgeMonths^2) + Sex, data = dat)
interact_plot(BPDiaAve_AgeMonthsSQ_Sex, pred = AgeMonths, modx = Sex,
plot.points = TRUE, interval = TRUE,
point.size = 0.7) +
ylab("Combined diastolic blood pressure") +
xlab("Age in Months")
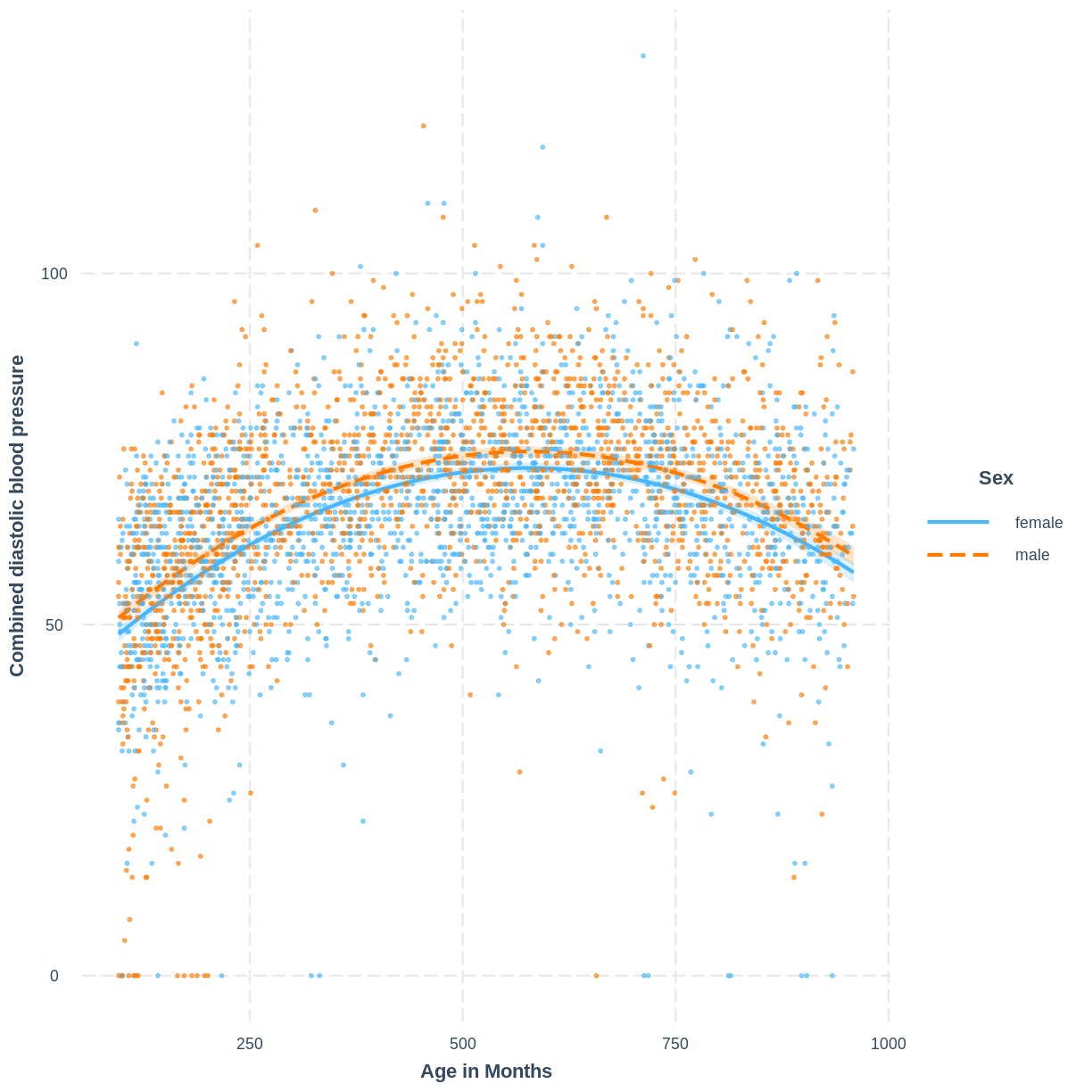
The non-linear relationship has now been modeled separately for levels of a categorical variable.
Recall that the additivity component of the linearity and additivity assumption means that the effect of any explanatory variable on the outcome variable does not depend on another explanatory variable in the model. In other words, that our model includes all necessary interactions between explanatory variables. In the above example, the additivity component does not appear to be violated: it does not look like the data requires an interaction between age and sex. However, to be sure of this we may compare the model fit of models with and without the interaction using the tools discussed in this episode.
Exercise
In the example above we saw that a model with a squared explanatory variable can also include separate intercepts for levels of a categorical variable.
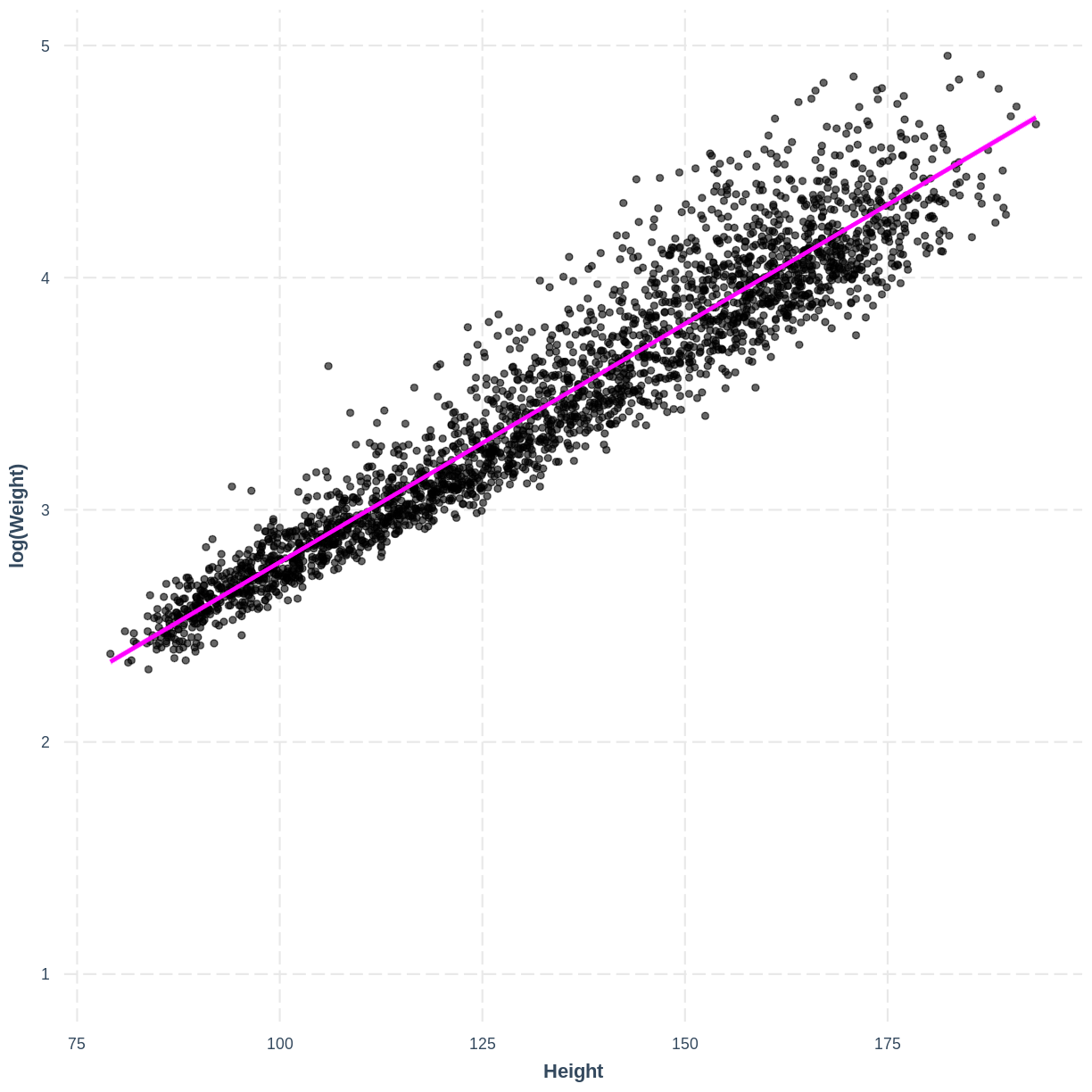
Recall our
child_logWeight_Height_lmmodel from the previous lesson, which modeled the relationship between the log of child weight and child height:child_logWeight_Height_lm <- dat %>% filter(Age < 18) %>% lm(formula = log(Weight) ~ Height) effect_plot(child_logWeight_Height_lm, pred = Height, plot.points = TRUE, interval = TRUE, line.colors = c("magenta"))
Extend this model by adding separate intercepts for the levels of the
Sexvariable. Visualise the model usinginteract_plot(). Do you think that including the interaction betweenHeightandSexis necessary?Solution
child_logWeight_Height_Sex <- dat %>% filter(Age < 18) %>% lm(formula = log(Weight) ~ Height + Sex) interact_plot(child_logWeight_Height_Sex, pred = Height, modx = Sex, plot.points = TRUE, interval = TRUE, point.size = 0.7)
There is little evidence from the plot that the effect of height on weight is different between sexes meaning that an interaction term is not needed. But we could assess the model fit with and without the interaction to check.
Independent errors
Recall that when observations in our data are grouped, this can result in a violation of the independent errors assumption. If there are a few levels in our grouping variable (say, less than 6) then we may overcome the violation by including the grouping variable as an explanatory variable in our model. If the grouping variable has many more levels, we may opt for a modelling approach more complex than multiple linear regression.
Exercise
In which of the following scenarios are we at risk of violating the independent errors assumption? In those cases, should we be working with an extra explanatory variable?
A) We are modelling the effect of average daily calorie intake on BMI in the adult UK population. We have one observation per participant and participants are known to belong to one of five income brackets.
B) We are modelling the effect of age on grip strength in adult females in the UK. Whether participants are physically active is known.
C) We are asking whether people’s sprinting speed is increased after partaking in an athletics course. Our data includes measurements of 100 participant’s sprinting speed before and after the course.Solution
A) Since we have multiple observations per income bracket, our observations are not independent. There are five levels in the income bracket variable, so we may chose to include income bracket as an explanatory variable.
B) Since we have multiple observations per level of physical activity, our observations are not independent. Since there are two levels in our physical activity variable, we could use physical activity as an explanatory variable in our model.
C) This data has two levels of grouping: within individuals (two measurements per individual) and timing (before/after the course). While timing can be included as an explanatory variable (two levels), it would not be appropriate to include individuals as an explanatory variable (100 levels). In this scenario we would opt for a more complex modelling approach (a mixed effect model).
Equal variance of errors (homoscedasticity)
This assumption states that the magnitude of variation in the residuals is not different across the fitted values or any explanatory variable. Importantly, when interactions are included in the model, the scale of the residuals should be checked at the interaction level. In the case of an interaction between continuous and categorical explanatory variables, this means colouring the points in the residuals vs explanatory variable plot by the levels of a categorical variable.
For example, below we create the diagnostic plots for our Hemoglobin_Age_Sex
model. We create plots of residuals vs. fitted (p1), residuals vs. age (p2)
and residuals vs. sex (p3). Notice that in the residuals vs. age plot, we colour
points by sex using colour = sex. This allows us to assess whether the residuals
are homogenously scattered across age, grouped by sex (i.e. at the interaction
level). Note that the / in p1 / p2 / p3 relies on the patchwork package
being loaded and results in the three graphs being plotted in vertical sequence.
residualData <- tibble(resid = resid(Hemoglobin_Age_Sex),
fitted = fitted(Hemoglobin_Age_Sex),
age = Hemoglobin_Age_Sex$model$Age,
sex = Hemoglobin_Age_Sex$model$Sex)
p1 <- ggplot(residualData, aes(x = fitted, y = resid)) +
geom_point(alpha = 0.3) +
geom_smooth() +
ylab("Residual") +
xlab("Fitted values")
p2 <- ggplot(residualData, aes(x = age, y = resid, colour = sex)) +
geom_point(alpha = 0.3) +
geom_smooth() +
ylab("Residual") +
xlab("Age")
p3 <- ggplot(residualData, aes(x = sex, y = resid)) +
geom_violin() +
geom_jitter(alpha = 0.3, width = 0.2) +
ylab("Residual") +
xlab("Sex")
p1 / p2 / p3

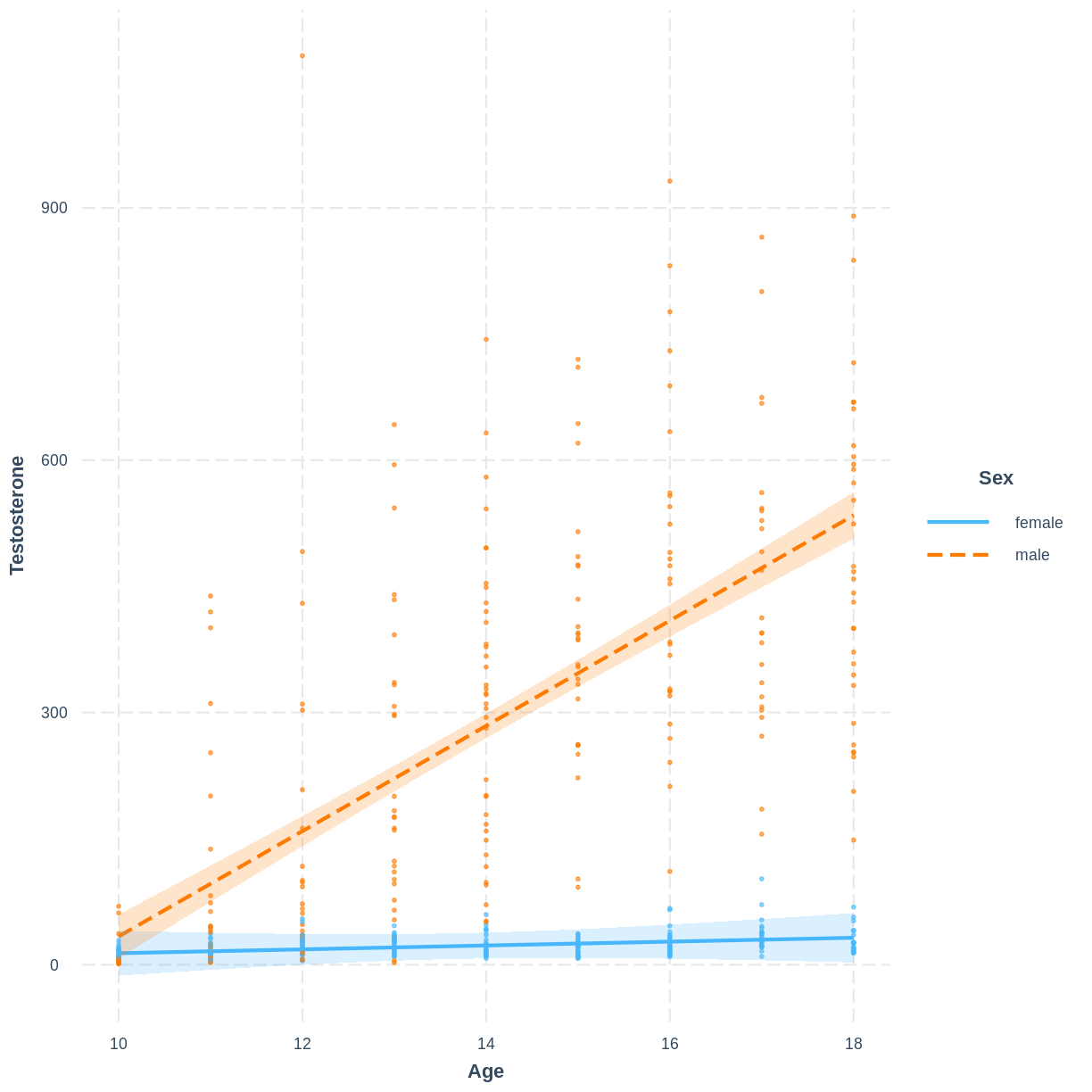
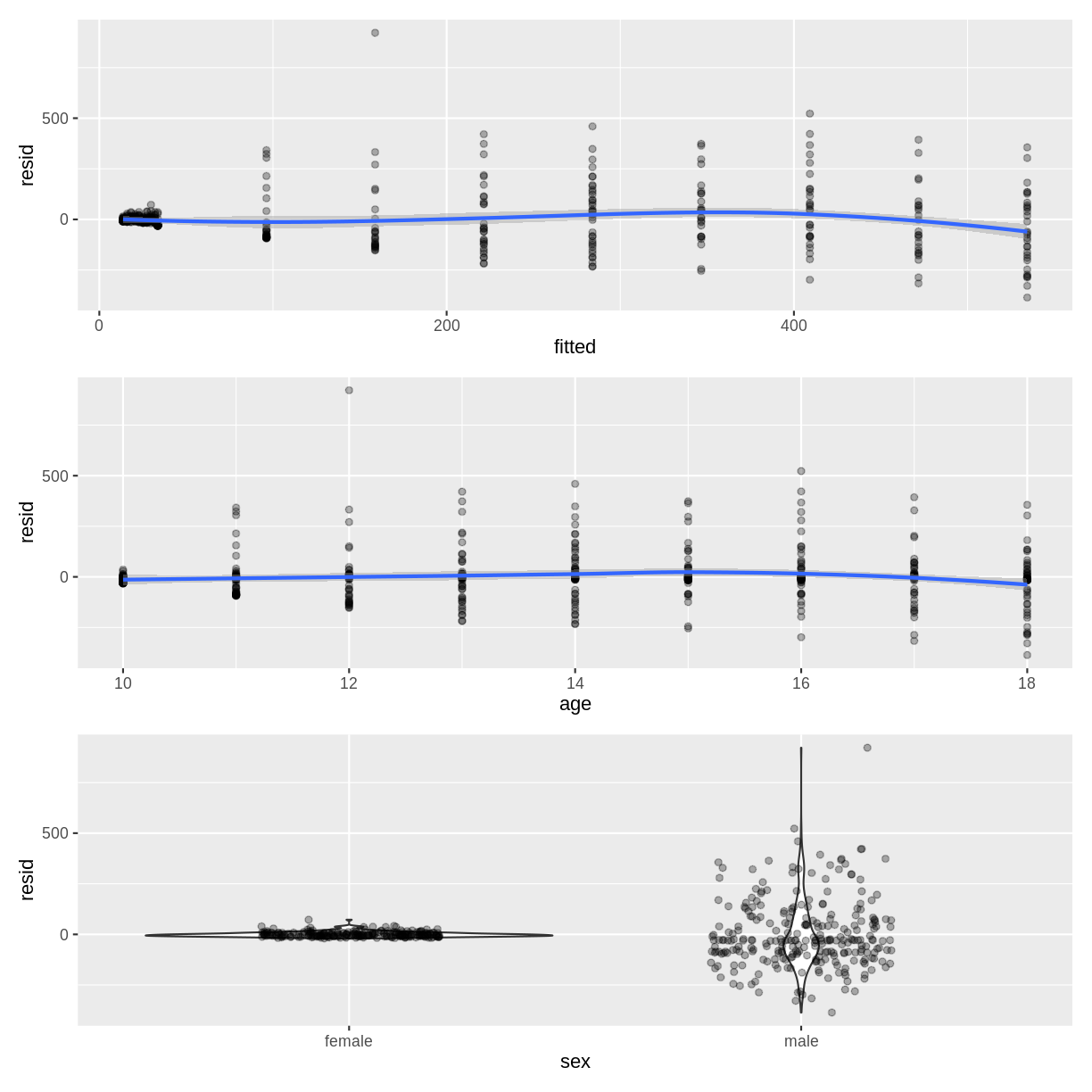
Exercise
A colleague is studying testosterone levels in children. They have fit a multiple linear regression model of testosterone as a function of age, sex and their interaction. The data and their model look as shown below:
This colleague approaches you for your thoughts on the following diagnostic plots, used to assess the homoscedasticity assumption.
A) What issues can you identify in the diagnostic plots?
B) How could the diagnostic plots be improved to be more informative?Solution
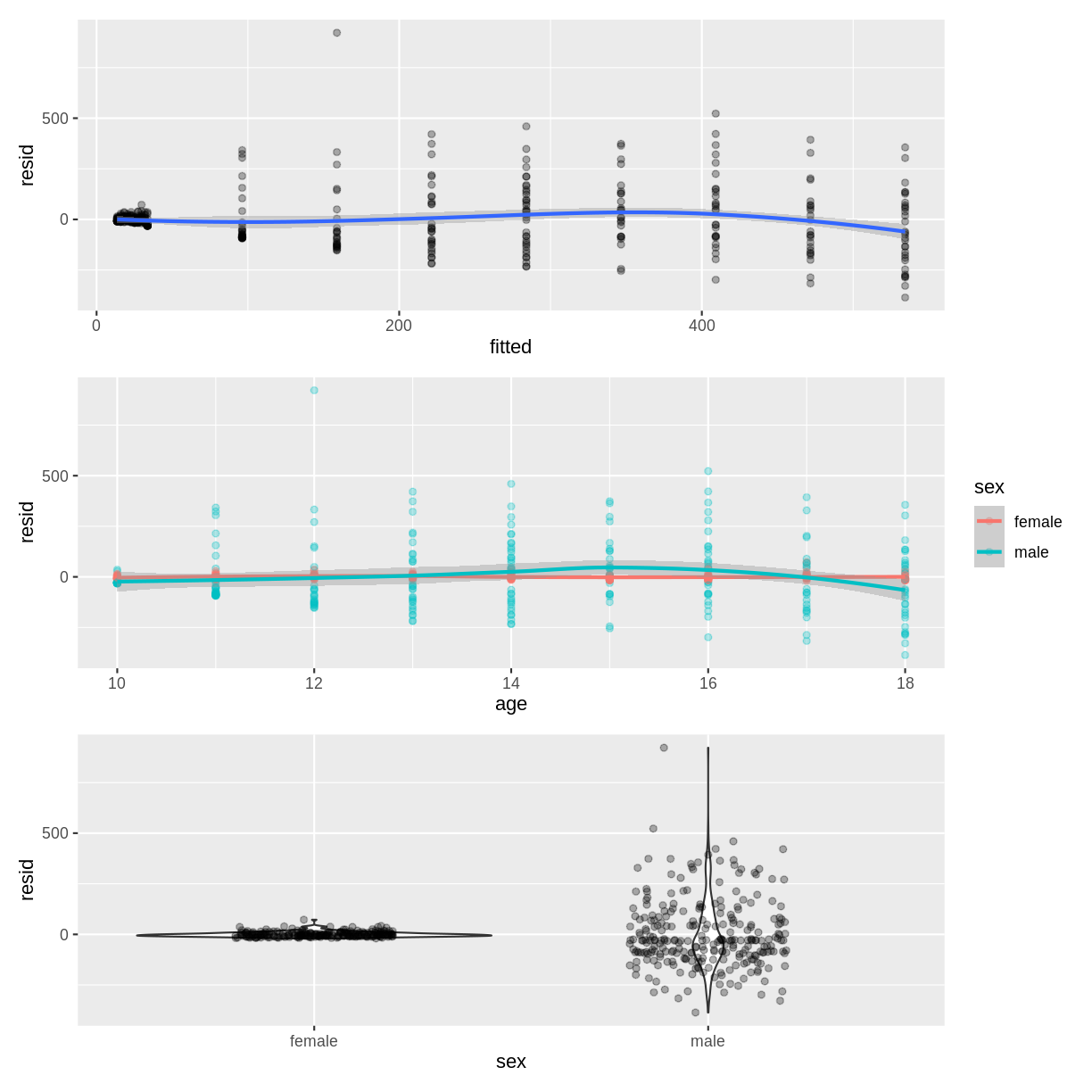
A) The residuals appear to fan out as the fitted values or age increase. The residuals also do not appear to be homogenous across sex, as the violin plot for males is much longer than for females.
B) The points in the scatterplot of age could be coloured by sex to assess the homoscedasticity assumption at the interaction level:
Normality of errors
Recall that this assumption states that the errors follow a normal distribution. When this assumption is strongly violated, predictions from the model are less reliable. Small deviations from normality may pose less of an issue. This assumption is assessed in the same way as for the case of the simple linear regression model, so we will not go through another exercise at this point.
Key Points
The adjusted R squared measure ensures that the metric does not increase simply due to the addition of a variable. The variable needs to improve model fit for the adjusted R squared to increase.
The same assumptions hold for simple and multiple linear regression, however more steps are involved in the assessment of the assumptions in the context of multiple linear regression.