Trial Design
Overview
Teaching: min
Exercises: minQuestions
What kind of on-farm experiments do we do?
How do we design these experiments efficiently?
Objectives
Know different types of common on-farm experiments
Import boundary file and AB line file
Create AB line file with code
utilize the functions to create simple trial designs
Trial design
To determine the most cost effective seed and fertilizer rates in a given field, we need to figure out:
- How much seed and nitrogen am I applying in this field now?
- How much does seed and nitrogen currently cost me?
- Should I apply more seed and/or nitrogen to improve profits?
- Could I apply less seed and/or nitrogen without reducing profits?
- How do I minimize the cost of inputs (seed and nitrogen) while maximizing yield (bushels per acre) at the current cost of seed and nitrogen?
In order to help figure this out, we create a grid over the field in which we combine areas of high, medium, and low rates of seed and fertilizer independently of each other. Some of the potential variations on this grid include:
- High seed and high fertilizer
- Low seed and high fertilizer
- Low seed and low fertilizer
- Several middle points between any of these combinations
Then we look at differences in yield data in the different areas of the field to determine:
- Whether more seed and more fertilizer made enough of a difference to be significant
- Where the most cost-effective seed and fertilizer rates are at the current prices of seed and nitrogen
For the next step, we will design our own experiments on the sample field. The only files we will need for the trial design are the boundary file and ab line.
Read and transform shape files
We will start by reading in the shape files we need:
boundary <- read_sf("data/boundary.gpkg") # read in boundary
abline <- read_sf("data/abline.gpkg") # read in AB line
Now let’s check the coordinate references of our two files:
st_crs(boundary)
Coordinate Reference System:
EPSG: 4326
proj4string: "+proj=longlat +datum=WGS84 +no_defs"
st_crs(abline)
Coordinate Reference System:
EPSG: 4326
proj4string: "+proj=longlat +datum=WGS84 +no_defs"
Since both of these are in lat/long and we want them in UTM, we’ll transform them:
trialarea <- st_transform_utm(boundary)
abline_utm <- st_transform_utm(abline)
Designing trials
We need decide on the details of experiment design before we get into any of the code. Relative parameters we need for the trial design include:
- plot dimensions
- number of treatments
- types of treatments, and
- treatment range.
Defining Parameters
In the following code, we are simply going to assign values to all the parameters that might be involved in the trial design. In this way, if we ever want to change any parameters, we can do it here, and need not to worry about the consistency for the whole code.
Now let’s design our grid with the following parameters:
width_in_meters <- 24 # width of grids is 24 meters
long_direction <- 'NS' # direction of grid that will be long in relation to AB line
short_direction <- 'EW' # direction of grid that will be short in relation to AB line
length_in_ft <- 180 # length of grids in feet
Make Grids
We’ll use our make_grids again function to generate this trial’s grid:
width <- m_to_ft(24) # convert meters to feet
design_grids_utm <- make_grids(trialarea, abline_utm,
long_in = long_direction,
short_in = short_direction,
length_ft = length_in_ft,
width_ft = width)
Next we want to make sure the coordinate reference frame of our trialarea is the same as our design_grids_utm grids and then take the intersection of these grids with our trial area as we did previously:
st_crs(design_grids_utm) <- st_crs(trialarea)
trial_grid <- st_intersection(trialarea, design_grids_utm)
Warning: attribute variables are assumed to be spatially constant throughout all
geometries
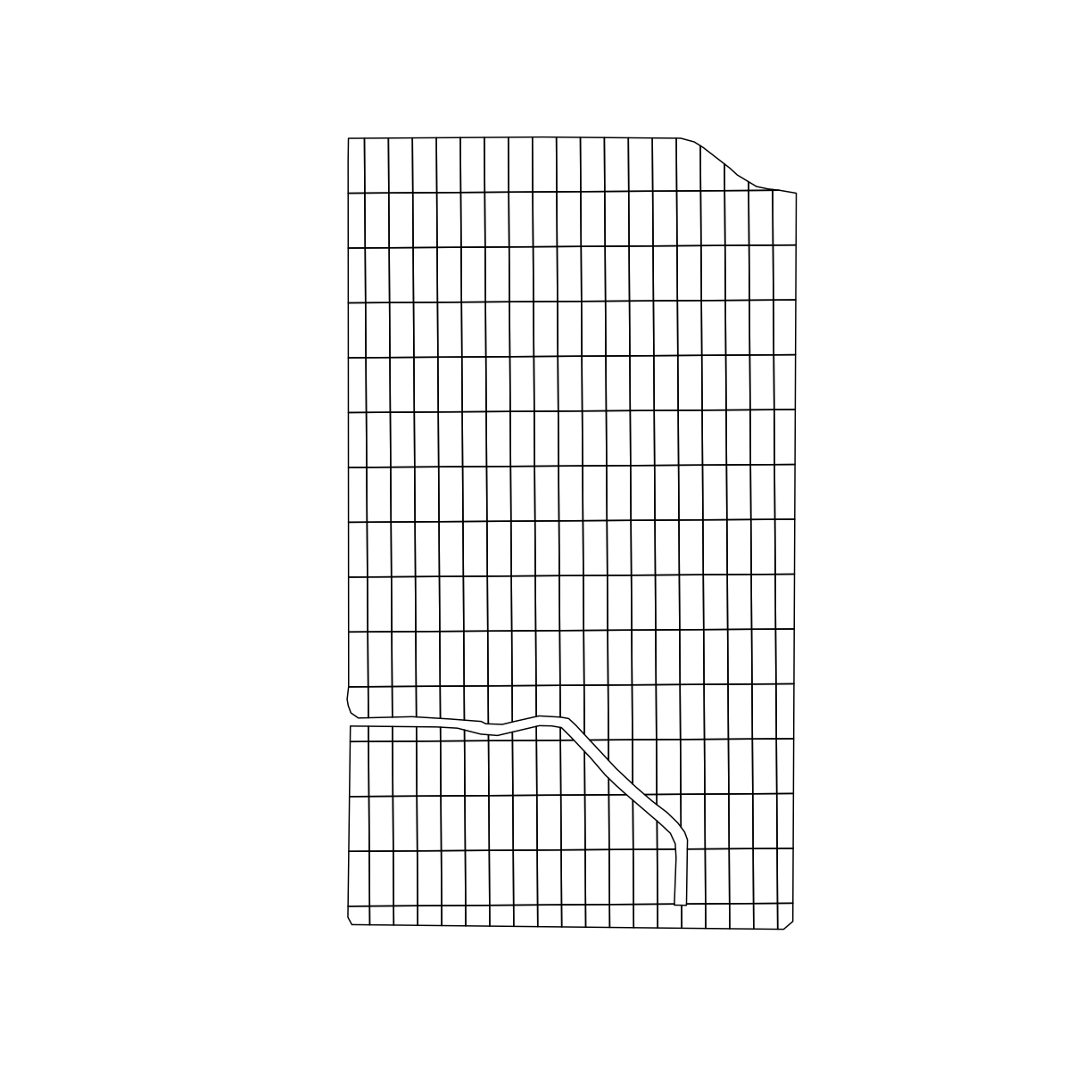
Let’s check out what our trial subplots look like:
plot(trial_grid$geom)
Determining subplot treatments
Now that we have the trial design plots, we need to assign different treatments to each plot. We can use the treat_assign function from functions.R to randomly assign seed rates and nitrogen rates to each plot on our grid.
We’ll select 4 different seed rates and 4 different nitrogen rates to deposit randomly on our grid:
seed_rates <- c(31000, 34000, 37000, 40000)
nitrogen_rates <- c(160,200,225,250)
The seed_quo and nitrogen_quo are the rates that will be applied to the headlands that are not part of the trial.
seed_quo <- 37000
nitrogen_quo <- 225
Lists of elements in R
You’ll see this definition of a list of numbers (or text) in R with a
c(). This is just a special way of saying all the elements in this list “belong” together, like with all of the numbers in a column of a spreadsheet “belonging” together.
Generating Treatment Map
We are now ready to generate our treatment plot:
whole_plot <- treat_assign(trialarea, trial_grid, head_buffer_ft = width,
seed_treat_rates = seed_rates,
nitrogen_treat_rates = nitrogen_rates,
seed_quo = seed_quo,
nitrogen_quo = nitrogen_quo,
set_seed=TRUE)
Simulating yields
Because you are generating your trial design “on the fly” in this workshop you will have different nitrogen and seed application rates than for the original dataset which measured the yields from a “real” trial. Because we want everybody to be using the same trial data we have set a flag
set_seed=TRUEin our call totreat_assign. When you do this on your own you can set this parameter toFALSEor just leave it out of the function call altogether.
Mapping Trials
Let’s look at what our trial looks like. First, as a shape file:
head(whole_plot)
Simple feature collection with 6 features and 4 fields
geometry type: POLYGON
dimension: XY
bbox: xmin: 342043.5 ymin: 4523203 xmax: 342066.9 ymax: 4523349
epsg (SRID): 32617
proj4string: +proj=utm +zone=17 +datum=WGS84 +units=m +no_defs
# A tibble: 6 x 5
id treat_type NRATE SEEDRATE geom
<chr> <dbl> <dbl> <dbl> <POLYGON [m]>
1 ID1 17 225 37000 ((342066.7 4523313, 342043.7 4523313, 342043.…
2 ID2 17 225 37000 ((342066.7 4523313, 342043.7 4523313, 342043.…
3 ID3 5 200 31000 ((342066.5 4523258, 342044.1 4523258, 342043.…
4 ID4 2 160 34000 ((342066.5 4523258, 342044.1 4523258, 342043.…
5 ID5 16 250 40000 ((342066.2 4523203, 342044.5 4523203, 342044.…
6 ID6 7 200 37000 ((342066.2 4523203, 342044.5 4523203, 342044.…
Or, we can look at the plots one at a time using a map_poly function from functions.R. For the seeding rate:
seed_plot <- map_poly(whole_plot, "SEEDRATE", "Seedrate Treatment")
seed_plot # to show our plot
 Here, we give
Here, we give map_poly our whole_plot geometry variable, what variable we want to show, in the above case, SEEDRATE, and the label to our legend, here Seedrate Treatement.
We can also plot the nitrogen application rate:
nitrogen_plot <- map_poly(whole_plot, "NRATE", "Nitrogen Treatment")
nitrogen_plot

We can also use the function tmap_arrange to show these plots side-by-side;
nitrogen_plot <- map_poly(whole_plot, "NRATE", "Nitrogen Treatment")
seed_plot <- map_poly(whole_plot, "SEEDRATE", "Seedrate Treatment")
treatment_plot_comp <- tmap_arrange(nitrogen_plot, seed_plot, ncol = 2, nrow = 1)
treatment_plot_comp
Key Points
Most of the code in this part would be using the functions, therefore understanding what different functions can be quite important
In designing the trials, the most important thing is to know how to design the experimental rates,and the tech part can be done by someone else