Introduction to scRNA-seq
Overview
Teaching: 10 min
Exercises: 5 minQuestions
What is the advantages of scRNA-seq?
What is the difference between bulk RNA-seq and scRNA-seq?
Objectives
Understand the the purpose of using next-generation sequencing (NGS)
Understand the pitfalls in bulk RNA-seq sequencing, and how can they be overcome
RNA-seq (RNA-sequencing) is a technique that can examine the quantity and sequences of RNA in a sample using next-generation sequencing (NGS). Compared to previous Sanger sequencing- and microarray-based methods, RNA-Seq provides far higher coverage and greater resolution of the dynamic nature of the transcriptome. Beyond quantifying gene expression, the data generated by RNA-Seq facilitate the discovery of novel transcripts, identification of alternatively spliced genes, and detection of allele-specific expression.
Note
We will refer to RNA-seq from now on by bulk RNA-seq to distinguish it from single-cell RNA-seq, which will be introduced later in this episode.
In order to perform bulk RNA-seq studies, samples have to be lysed, and the RNA is extracted. The following steps involve converting a sample of RNA to a cDNA library, which is then sequenced and mapped against a reference genome. The original goal of RNA sequencing was to compare the actual transcript abundance between samples.
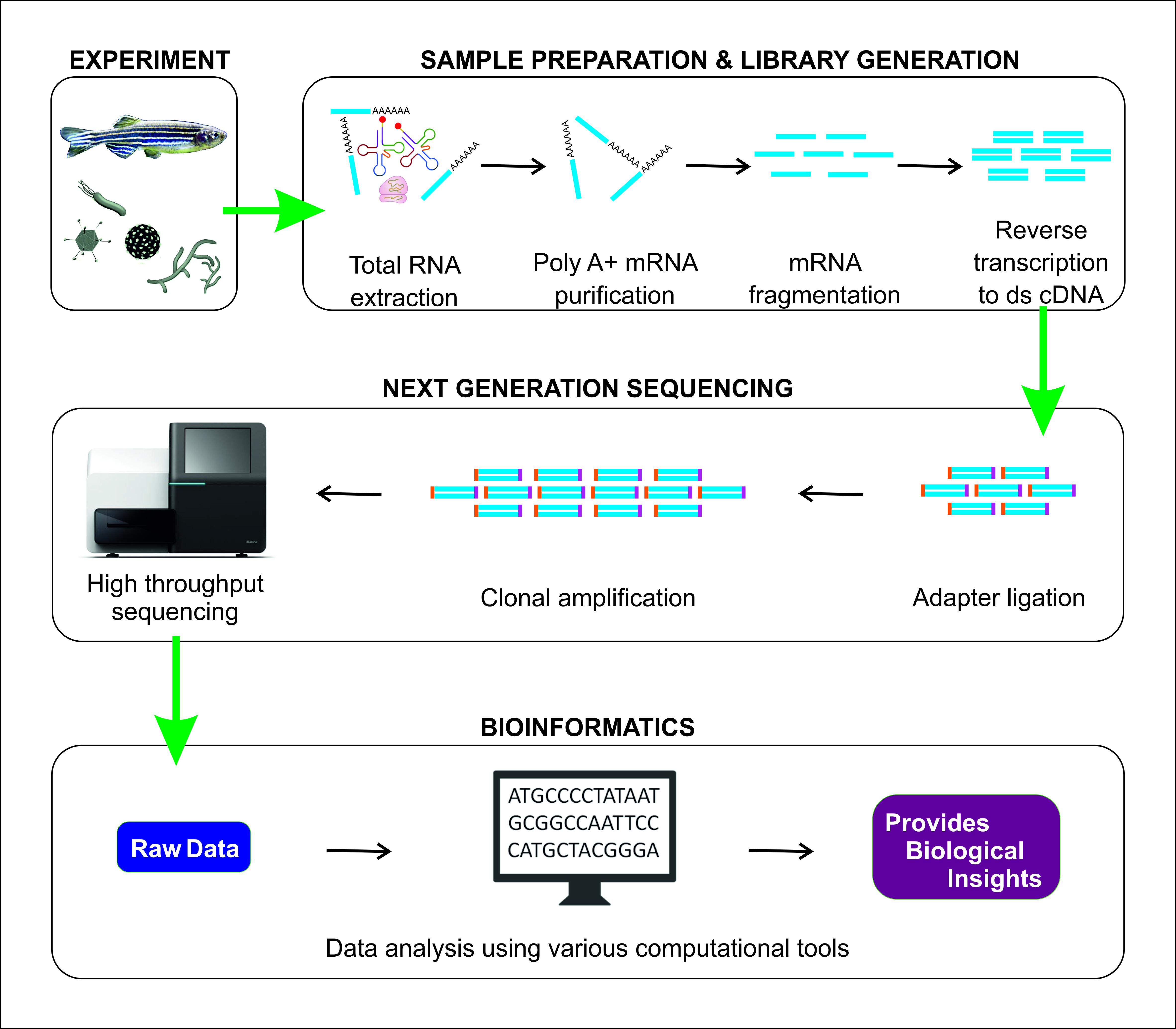
Image is taken from “Sudhagar, A.; Kumar, G.; El-Matbouli, M. Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review. Int. J. Mol. Sci. 2018, 19, 245. https://doi.org/10.3390/ijms19010245” under CC-BY license
Question
What is a cDNA library prepration, and why it is used in the bulk RNA-seq?
Solution
RNA is reverse transcribed to cDNA because DNA is more stable and to allow for amplification (which uses DNA polymerases) and leverage more mature DNA sequencing technology. While direct sequencing of RNA molecules is possible, most RNA-Seq experiments are carried out on instruments that sequence DNA molecules due to the technical maturity of commercial instruments designed for DNA-based sequencing. Therefore, cDNA library preparation from RNA is a required step for RNA-Seq. Each cDNA in an RNA-Seq library is composed of a cDNA insert of certain size flanked by adapter sequences, as required for amplification and sequencing on a specific platform.
Pitfall of bulk RNA-seq
- Bulk RNA-seq only provide the average measurement for all the cells. It does not provide insights into the stochastic nature of gene expression. The image below show that although average can be the same, the appearance of the dataset can be vary dramatically which means that average can be misleading and meaningless.
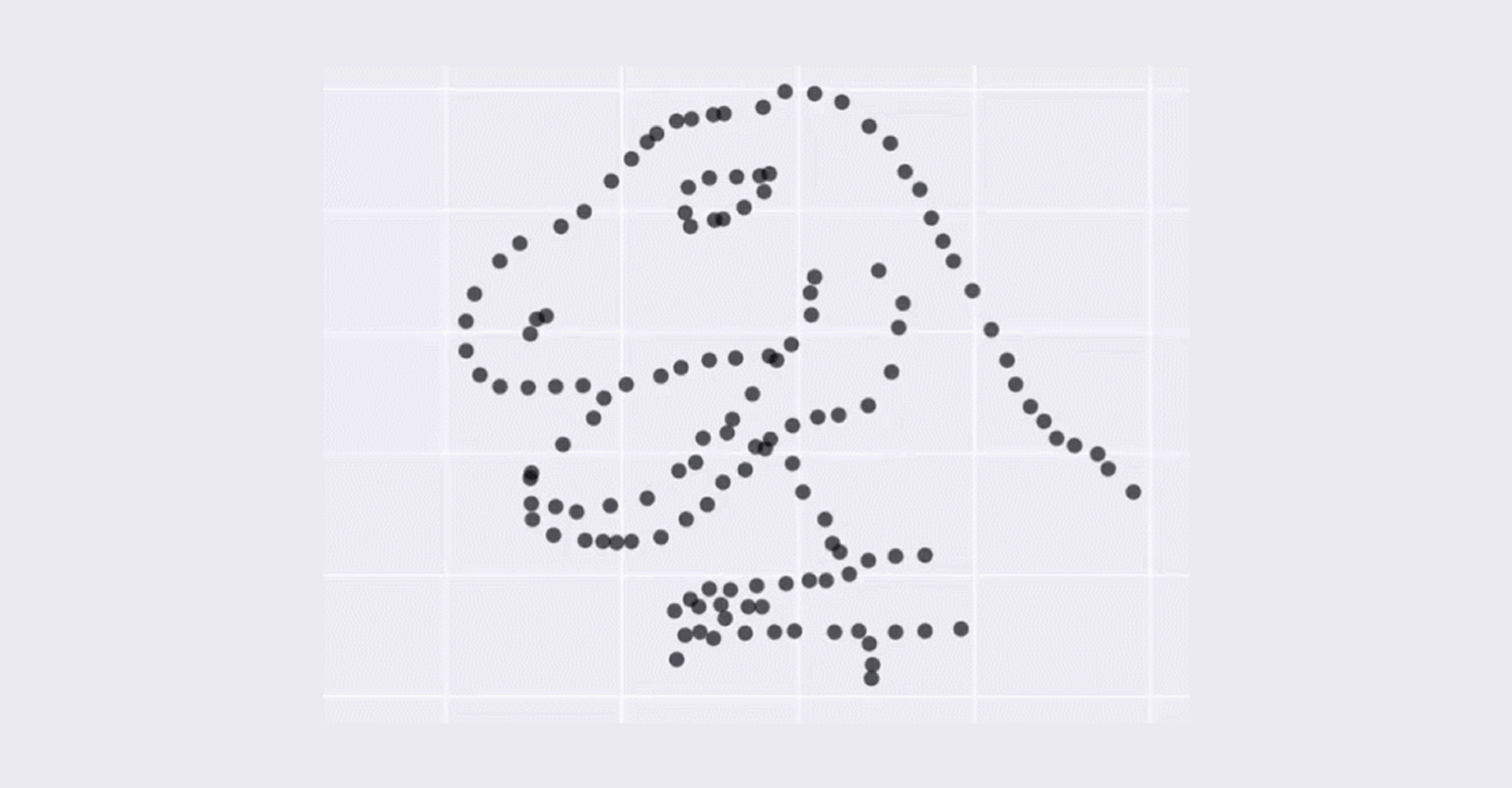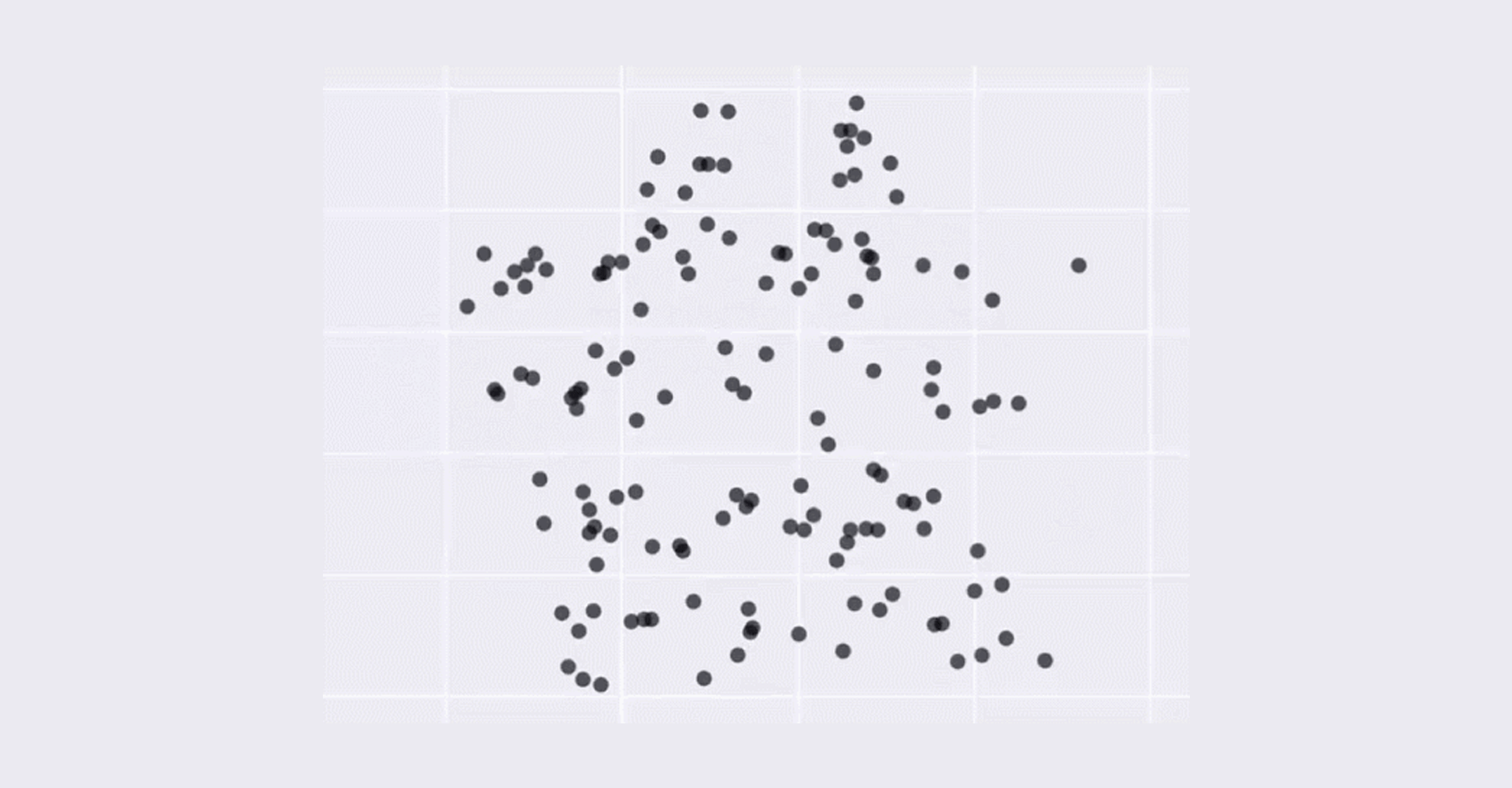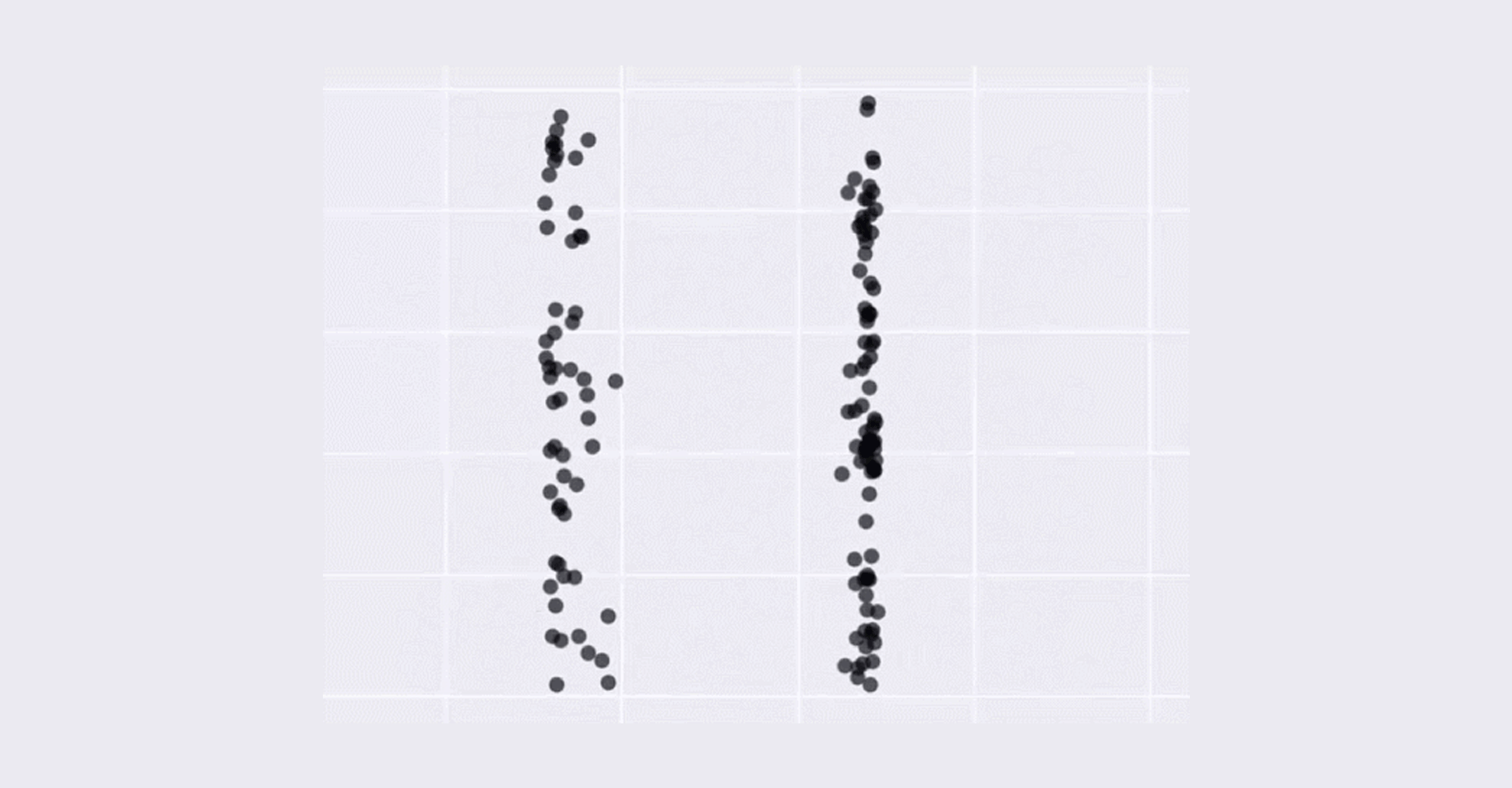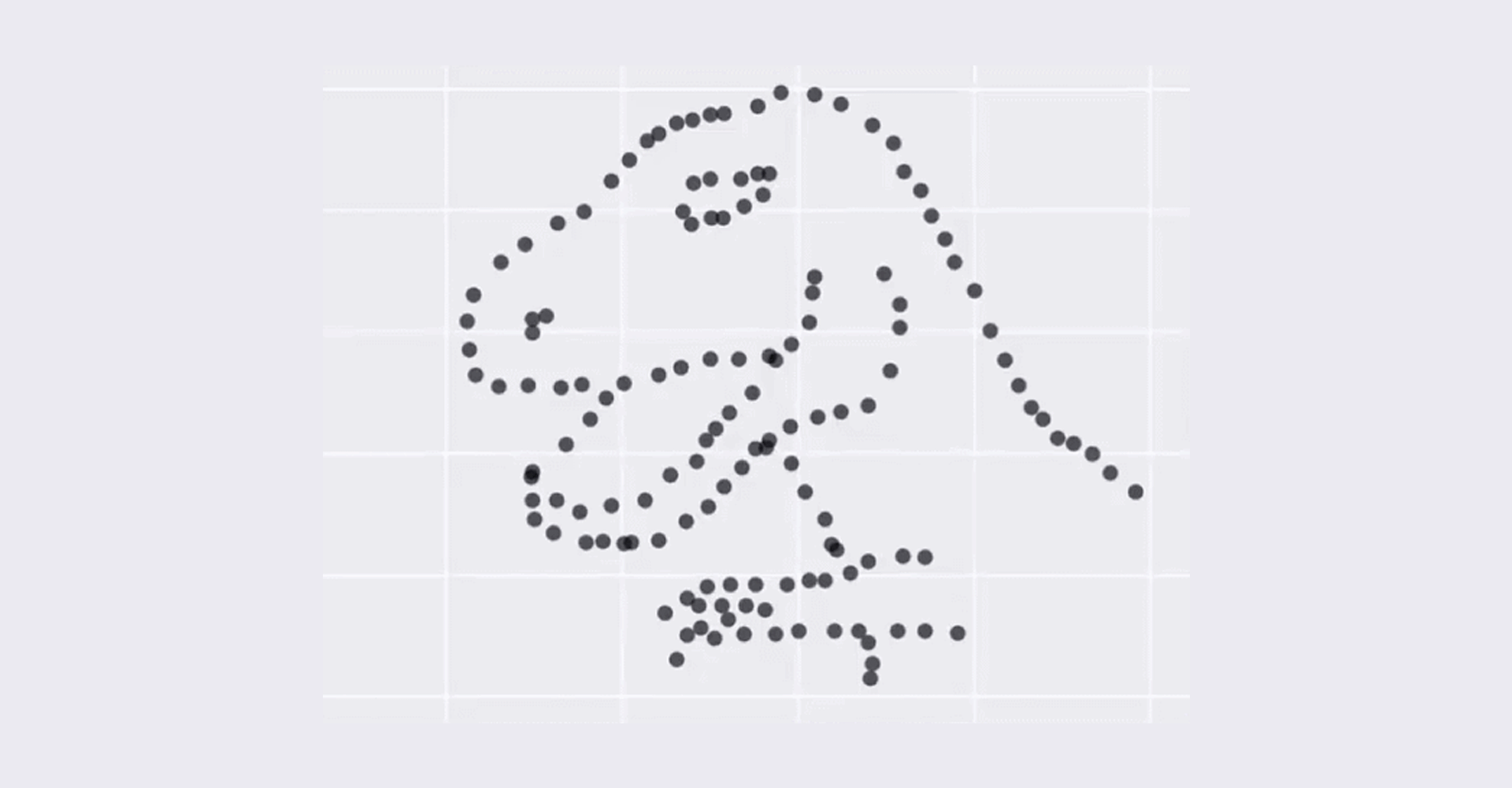
This dataset above has always the same mean (X Mean:54.26, Y Mean: 47.83, X SD:16.76, Y SD:26.93, Corr:-0.06) but varied appearance - Taken from Justin Matejka and George Fitzmaurice (Autodesk Research, Toronto)
- Insufficient for studying heterogeneous systems, e.g. early development studies, complex tissues (brain)
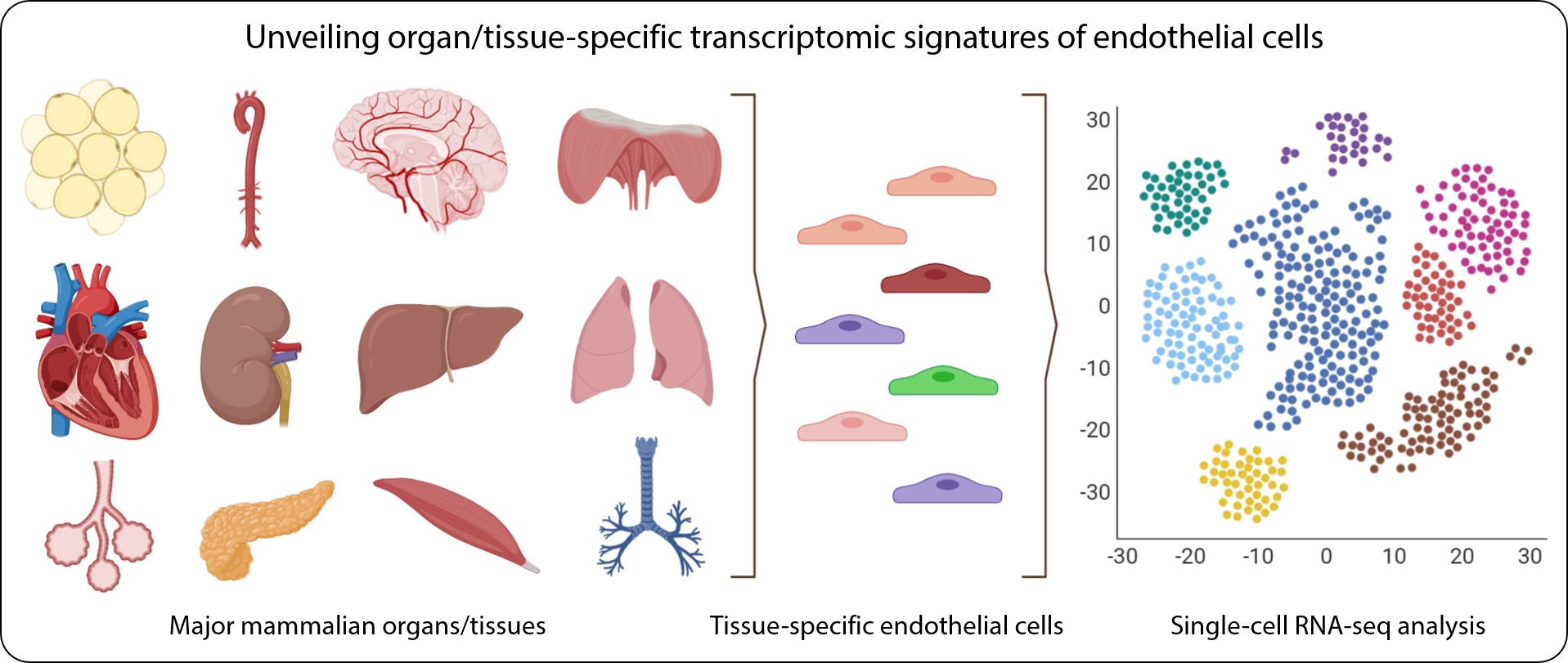
Image is taken from https://doi.org/10.1161/CIRCULATIONAHA.119.041433 under CC-BY license
Therefore, scRNA-seq was introduced to overcome the limitations of bulk RNA-seq.
Question
What do we mean by “stochastic nature of gene expression”?
Solution
It means that there are fluctuations in the abundance of expressed genes at the single-cell level because of the heterogeneity within populations of genetically identical cells.
Kærn, M., Elston, T., Blake, W. et al. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6, 451–464 (2005). https://doi.org/10.1038/nrg1615
- Even Though cell lines might seem more homogenous than tissue. They undergoes cell cycle heterogeneity, which can’t be taken into account during bulk RNA-seq
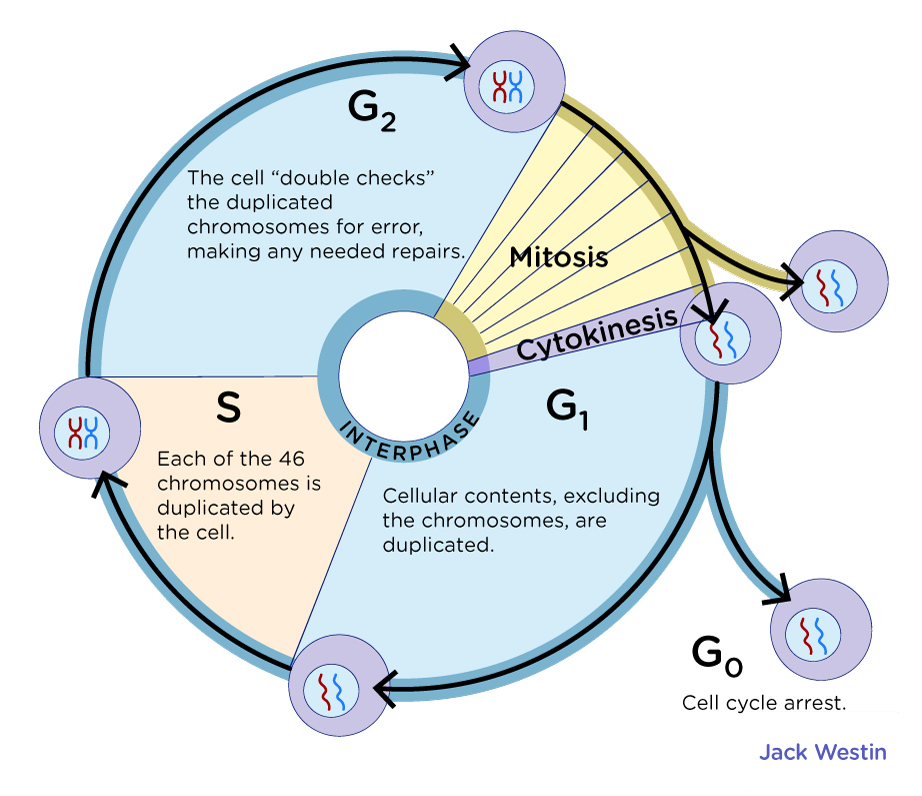
Image is taken from jackwestin.com under CC-BY license
Introduction to single-cell RNA-seq
The main difference between bulk and single-cell RNA-seq (scRNA-seq) is that each sequencing library represents a single cell, instead of a population of cells. Bulk sample analysis is just like putting a fruit salad into a blender - the taste is an average of all ingredients. Analyzing single cells is like tasting each individual piece of fruit to gain a much more nuanced understanding of the composition of the fruit salad Single cell genomics
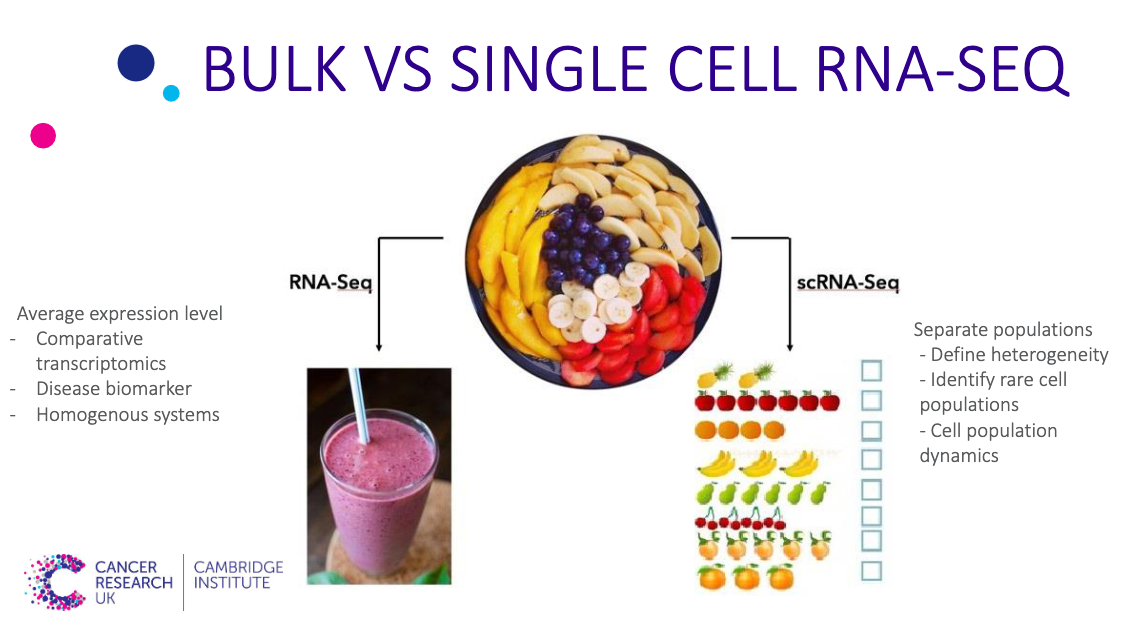
The advantages of using scRNA-seq
- It allows the measurement of the distribution of expression levels for each gene across a population of cells.
- It also allows to study new biological questions in which cell-specific changes in transcriptome are important, e.g. cell type identification, heterogeneity of cell responses, stochasticity of gene expression, inference of gene regulatory networks across the cells.
- The main advantage of scRNA-seq is that the cellular resolution and the genome-wide scope make it possible to address intractable issues using other methods, e.g. bulk RNA-seq or single-cell RT-qPCR.
However, to analyse scRNA-seq data, novel methods are required, and some of the underlying assumptions for the methods developed for bulk RNA-seq experiments are no longer valid. There are also many different protocols in use, e.g. SMART-seq2 (Picelli et al. 2013), CELL-seq (Hashimshony et al. 2012) and Drop-seq (Macosko et al. 2015), which adds to the complexity of analysing scRNA-seq. In the next episode, we will briefly explore the various experimental methods currently used for scRNA-seq.
Resources Consulted & Recommended Reading
- Mehmet Tekman, Hans-Rudolf Hotz, Daniel Blankenberg, Wendi Bacon, 2021 Pre-processing of 10X Single-Cell RNA Datasets (Galaxy Training Materials). /training-material/topics/transcriptomics/tutorials/scrna-preprocessing-tenx/tutorial.html Online; accessed Tue Apr 06 2021
- Batut et al., 2018 Community-Driven Data Analysis Training for Biology Cell Systems 10.1016/j.cels.2018.05.012
Key Points
scRNA-seq offers many advantages over bulk RNA-seq
scRNA-seq requires more pre-processing than bulk RNA-seq