Diagnosing Issues and Improving Robustness
Overview
Teaching: 30 min
Exercises: 20 minQuestions
Once we know our program has errors, how can we locate them in the code?
How can we make our programs more resilient to failure?
Objectives
Use a debugger to explore behaviour of a running program
Describe and identify edge and corner test cases and explain why they are important
Apply error handling and defensive programming techniques to improve robustness of a program
Integrate linting tool style checking into a continuous integration job
Introduction
Unit testing can tell us something is wrong in our code and give a rough idea of where the error is by which test(s) are failing. But it does not tell us exactly where the problem is (i.e. what line of code), or how it came about. To give us a better idea of what is going on, we can:
- output program state at various points, e.g. by using print statements to output the contents of variables,
- use a logging capability to output the state of everything as the program progresses, or
- look at intermediately generated files.
But such approaches are often time consuming and sometimes not enough to fully pinpoint the issue. In complex programs, like simulation codes, we often need to get inside the code while it is running and explore. This is where using a debugger can be useful.
Setting the Scene
Let us add a new function called data_normalise() to our catchment example
to normalise a given measurement data array so that all entries fall between 0 and 1.
(Make sure you create a new feature branch for this work off your develop branch.)
To normalise each set of measurement data
we need to divide it by the maximum measurement value taken.
To do so, we can add the following code to catchment/models.py:
import numpy as np
...
def data_normalise(data):
"""Normalise any given 2D data array"""
max = np.array(np.max(data, axis=1))
return data / max[np.newaxis, :]
Note: there are intentional mistakes in the above code, which will be detected by further testing and code style checking below so bear with us for the moment.
For this work we will make use of the NumPy library. Pandas dataframes are built on top of NumPy arrays, which means that we can make use of the NumPy toolkit for manipulating Pandas data if we find that this would be more appropriate than using a Pandas tool.
In the code above, we first go column by column
and find the maximum data value for each measurement site
and store these values in a 1-dimensional NumPy array max.
We then want to use NumPy’s element-wise division,
to divide each value in every column of measurement data
(belonging to the same site)
by the maximum value for that site stored in the 1D array max.
However, we cannot do that division automatically
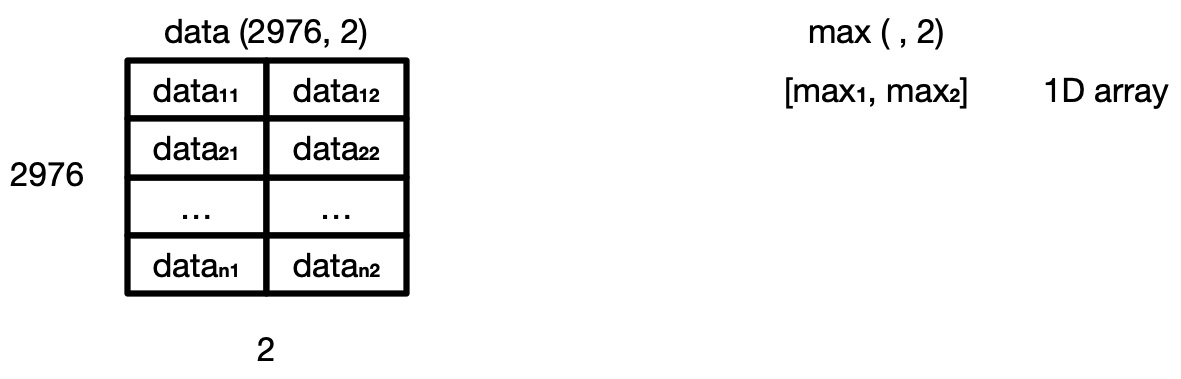
as data is a 2D array (of shape (2976, 2))
and max is a 1D array (of shape (, 2)),
which means that their shapes are not compatible.
Hence, to make sure that we can perform this division and get the expected result,
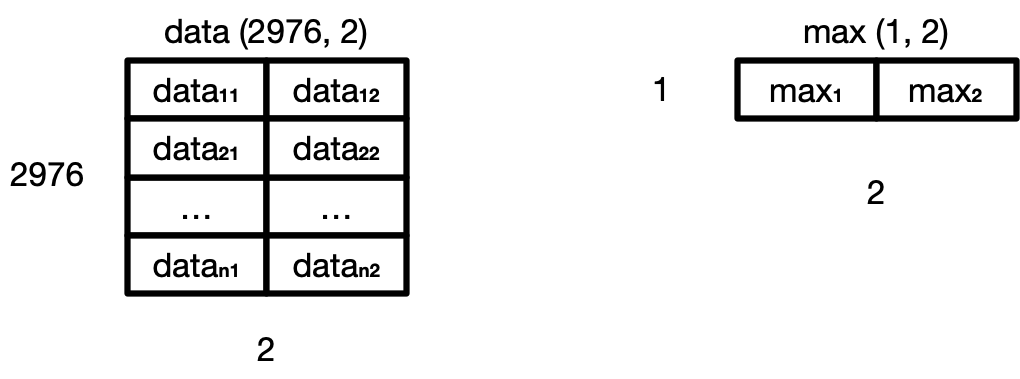
we need to convert max to be a 2D array
by using the newaxis index operator to insert a new axis into max,
making it a 2D array of shape (1, 2).
Now the division will give us the expected result.
Even though the shapes are not identical,
NumPy’s automatic broadcasting (adjustment of shapes) will make sure that
the shape of the 2D max array is now “stretched” (“broadcast”)
to match that of data - i.e. (2976, 2),
and element-wise division can be performed.
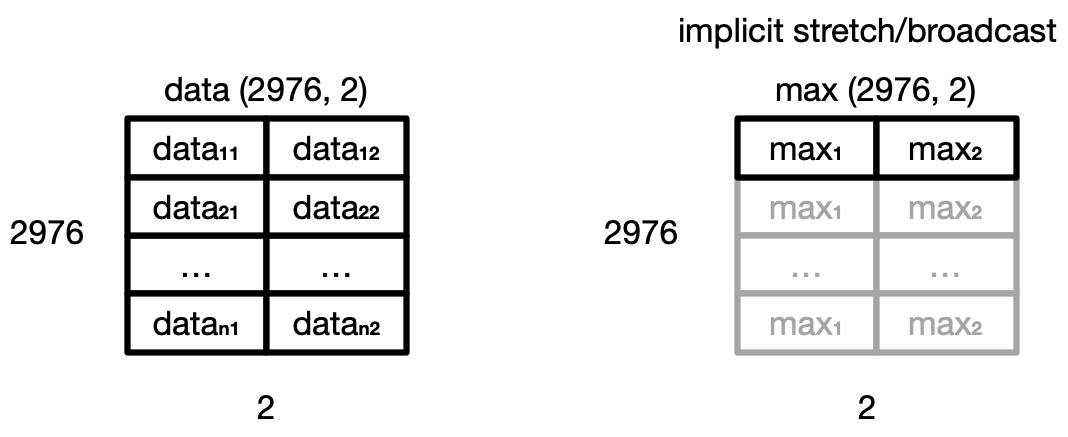
Broadcasting
The term broadcasting describes how NumPy treats arrays with different shapes during arithmetic operations. Subject to certain constraints, the smaller array is “broadcast” across the larger array so that they have compatible shapes. Be careful, though, to understand how the arrays get stretched to avoid getting unexpected results.
Note there is an assumption in this calculation that the minimum value we want is always zero. This is a sensible assumption for this particular application, since the zero value is a special case indicating that a patient experienced no inflammation on a particular day.
Let us now add a new test in tests/test_models.py
to check that the normalisation function is correct for some test data.
@pytest.mark.parametrize(
"test_data, test_index, test_columns, expected_data, expected_index, expected_columns",
[
(
[[1, 2, 3], [4, 5, 6], [7, 8, 9]],
[pd.to_datetime('2000-01-01 01:00'),
pd.to_datetime('2000-01-01 02:00'),
pd.to_datetime('2000-01-01 03:00')],
['A', 'B', 'C'],
[[0.14, 0.25, 0.33], [0.57, 0.63, 0.66], [1.0, 1.0, 1.0]],
[pd.to_datetime('2000-01-01 01:00'),
pd.to_datetime('2000-01-01 02:00'),
pd.to_datetime('2000-01-01 03:00')],
['A', 'B', 'C']
),
])
def test_normalise(test_data, test_index, test_columns, expected_data, expected_index, expected_columns):
"""Test normalisation works for arrays of one and positive integers.
Assumption that test accuracy of two decimal places is sufficient."""
from catchment.models import data_normalise
pdt.assert_frame_equal(data_normalise(pd.DataFrame(data=test_data, index=test_index, columns=test_columns)),
pd.DataFrame(data=expected_data, index=expected_index, columns=expected_columns),
atol=1e-2)
Note another assumption made here that a test accuracy of two decimal places is sufficient -
so we state this explicitly by setting the absolute tolerance of the tests using atol=1e-2,
and have rounded our expected values up accordingly.
The assert_frame_equal Pandas testing function allows
the setting of absolute (atol) and relative (rtol) tolerances
to enable testing against values that are almost equal:
very useful when we have numbers with arbitrary decimal places
and are only concerned with a certain degree of precision,
like the test case above.
Run the tests again using python -m pytest tests/test_models.py
and you will note that the new test is failing,
with an error message that does not give many clues as to what went wrong.
tests/test_models.py:142:
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
pandas/_libs/testing.pyx:52: in pandas._libs.testing.assert_almost_equal
???
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
> ???
E AssertionError: DataFrame.iloc[:, 0] (column name="A") are different
E
E DataFrame.iloc[:, 0] (column name="A") values are different (100.0 %)
E [index]: [2000-01-01T01:00:00.000000000, 2000-01-01T02:00:00.000000000, 2000-01-01T03:00:00.000000000]
E [left]: [0.3333333333333333, 1.3333333333333333, 2.3333333333333335]
E [right]: [0.14, 0.57, 1.0]
Let us use a debugger at this point to see what is going on and why the function failed.
Debugging in VS Code
Think of debugging like performing exploratory surgery - on code! Debuggers allow us to peer at the internal workings of a program, such as variables and other state, as it performs its functions.
Running Tests Within VS Code
Firstly, to make it easier to track what’s going on, we can set up VS Code to run and debug our tests instead of running them from the command line. If you have not done so already, you will first need to enable the Pytest framework in VS Code. You can do this by:
- In VS Code, select the ‘Testing’ tab on the Activity Bar on the left side of the window
(icon resembles a chemistry flask/beaker).
If you have not yet configured any tests,
you will see a blue
Configure Python Testsbutton. If tests have already been configured and are incorrect, or you wish to review this process, open the Command Palette (Command+Shift+P for Mac, Control+Shift+P for Windows) and search forPython: Configure Tests(keeping the>character at the start of the search string). - Then, in the text search box that appears at the top of the edit window,
type
pytestand selectpytest pytest frameworkfrom the drop-down list. - You will be asked for the root directory of your tests.
Select the
testsfolder in our project folder. - The left hand panel will then display the
testsfolder with each of the files it contains.
We can now run pytest over our tests in VS Code,
similarly to how we ran our catchment-analysis.py script before.
In the Testing panel, right-click the test_models.py file in the tests folder.
under the tests directory in the file navigation window on the left,
and select Run test.
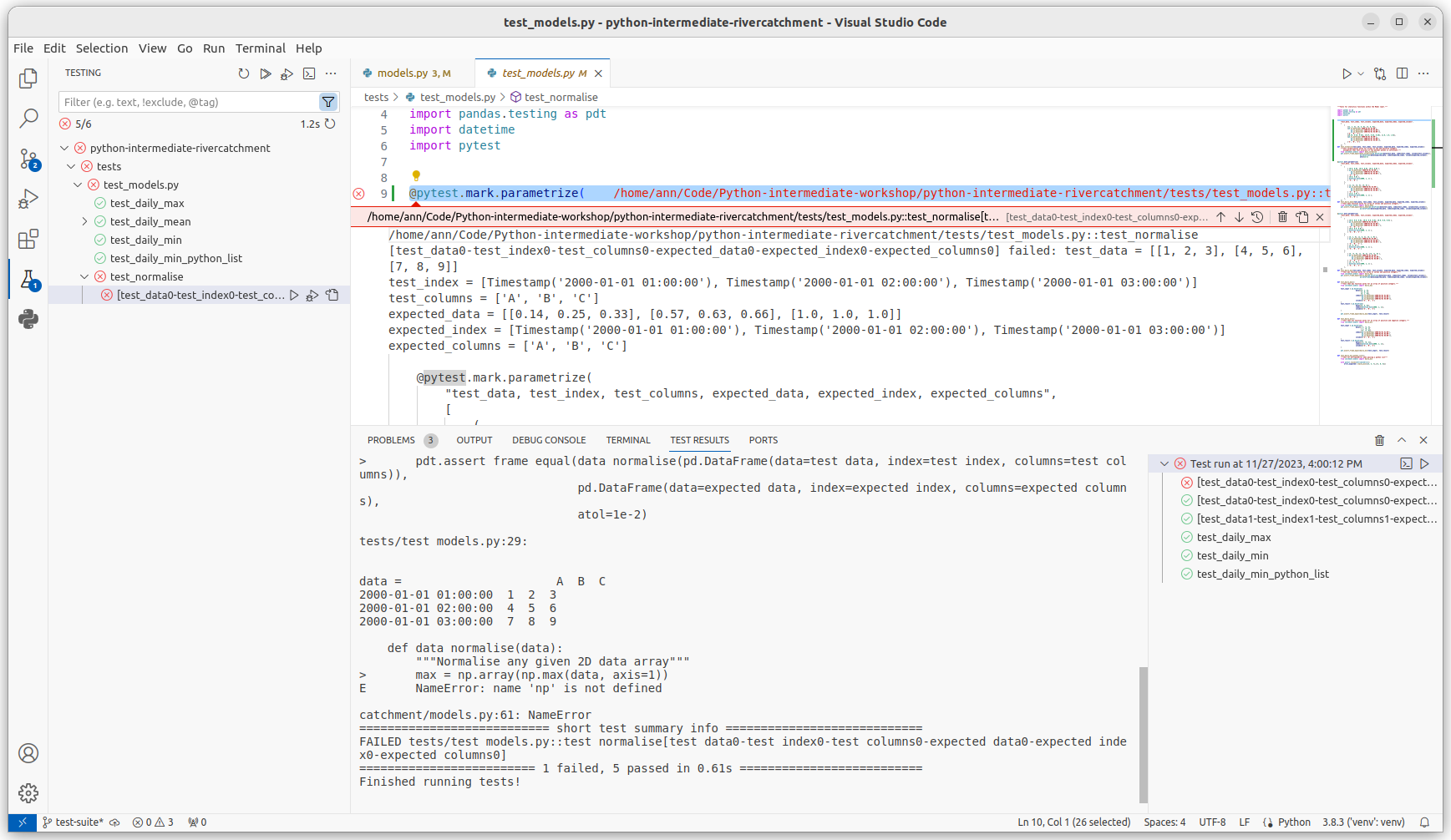
You’ll see the results of the tests appear in VS Code in a bottom panel.
If you scroll down in that panel you should see
the failed test_normalise() test result
looking something like the following:
We can also run our test functions individually.
Click on a green check next to a test function
in our test_models.py script in VS Code,
(or right click it and select Run test),
we can run just that test:
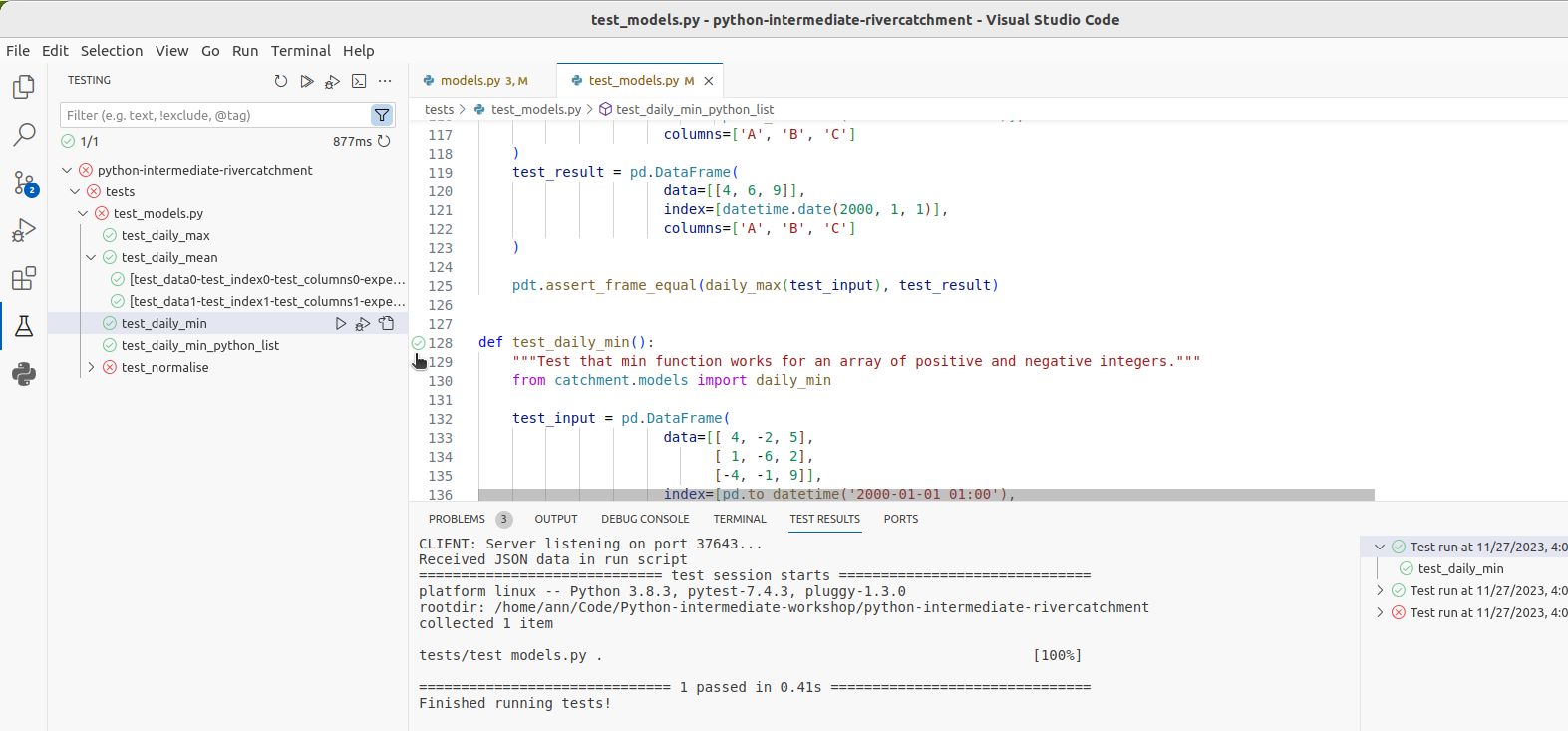
Click on the Run Test button next to test_normalise,
and you will be able to see that VS Code runs just that test function,
and we see the same AssertionError that we saw before.
Running the Debugger
Now we want to use the debugger to investigate
what is happening inside the data_normalise function.
To do this we will add a breakpoint in the code.
A breakpoint will pause execution at that point allowing us to explore the state of the program.
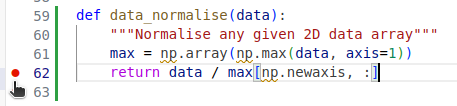
To set a breakpoint, navigate to the models.py file
and move your mouse to the return statement of the data_normalise function.
Click on just to the right of the line number for that line
and a small red dot will appear,
indicating that you have placed a breakpoint on that line.
Now if you find test_models.py in the Testing panel, and locate the green play/right-arrow marker
for the test_normalise function
(in VS Code this appears next to the decorator function
@pytest.mark.parameterize that we recently added to test_normalise).
Right click on that arrow and select Debug Test from the drop down menu.
You will notice that execution will be paused
at the return statement of data_normalise,
where we placed our breakpoint.
In the debug panel that appears below,
we can now investigate the exact state of the program
prior to it executing this line of code.
In the debug panel on the left hand side, you will be able to see three sections that looks something like the following:
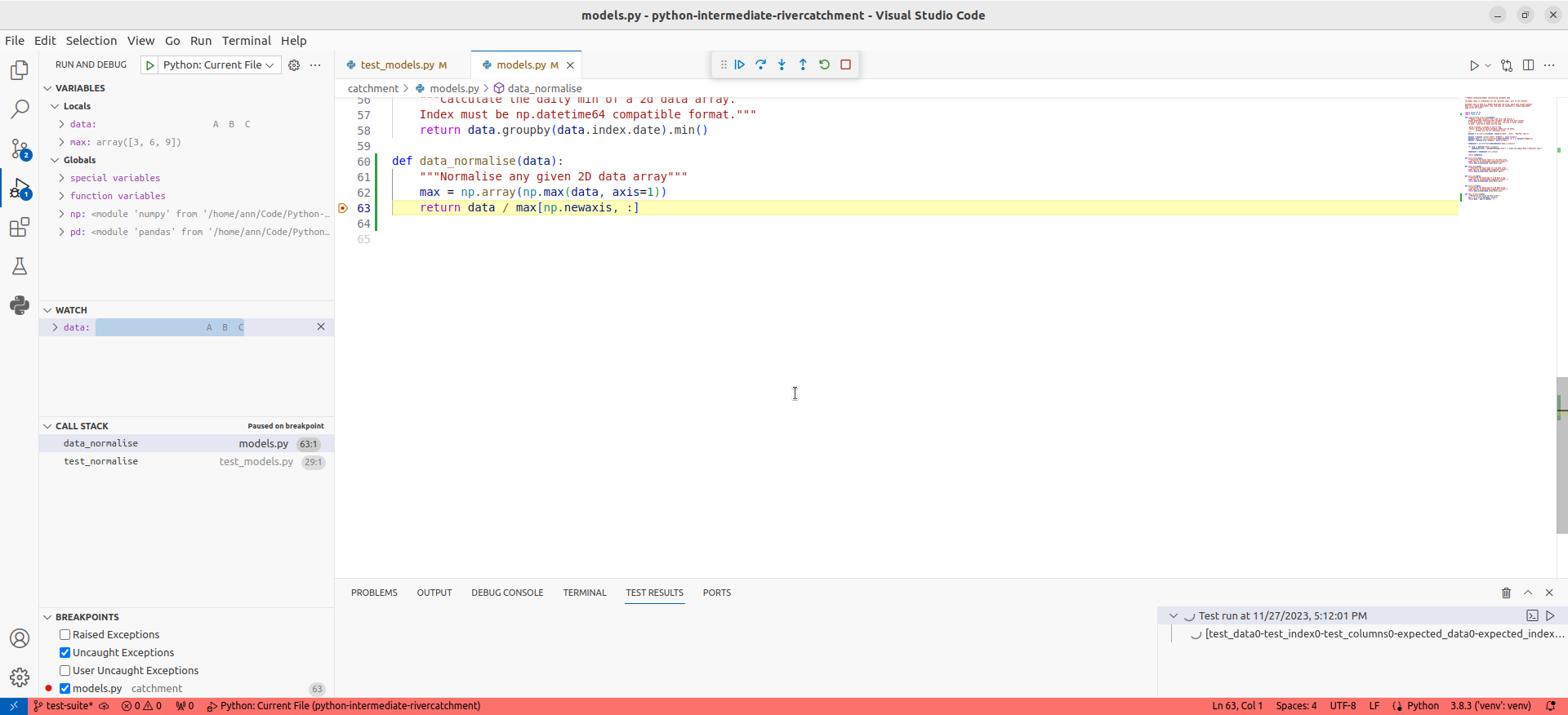
- The
Variablessection at the top, which displays the local and global variables currently in memory. You will be able to see thedataarray that is input to thedata_normalisefunction, as well as themaxlocal array that was created to hold the maximum inflammation values for each patient. - The
Watchsection in the middle where we can add/remove expressions that we need to evaluate. - The
Call Stacksection at the bottom, which shows the chain of functions that have been executed to lead to this point. We can traverse this chain of functions if we wish, to observe the state of each function.
We also have the ability run any Python code we wish at this point to explore the state of the program even further! This is useful if you want to view a particular combination of variables, or perhaps a single element or slice of an array to see what went wrong.
Select the Debug Console tab in the bottom panel,
and you’ll be presented with a Python prompt.
Try putting in the expression max[np.newaxis, :] into the console,
and you will be able to see the row vector that we are dividing data by
in the return line of the function.
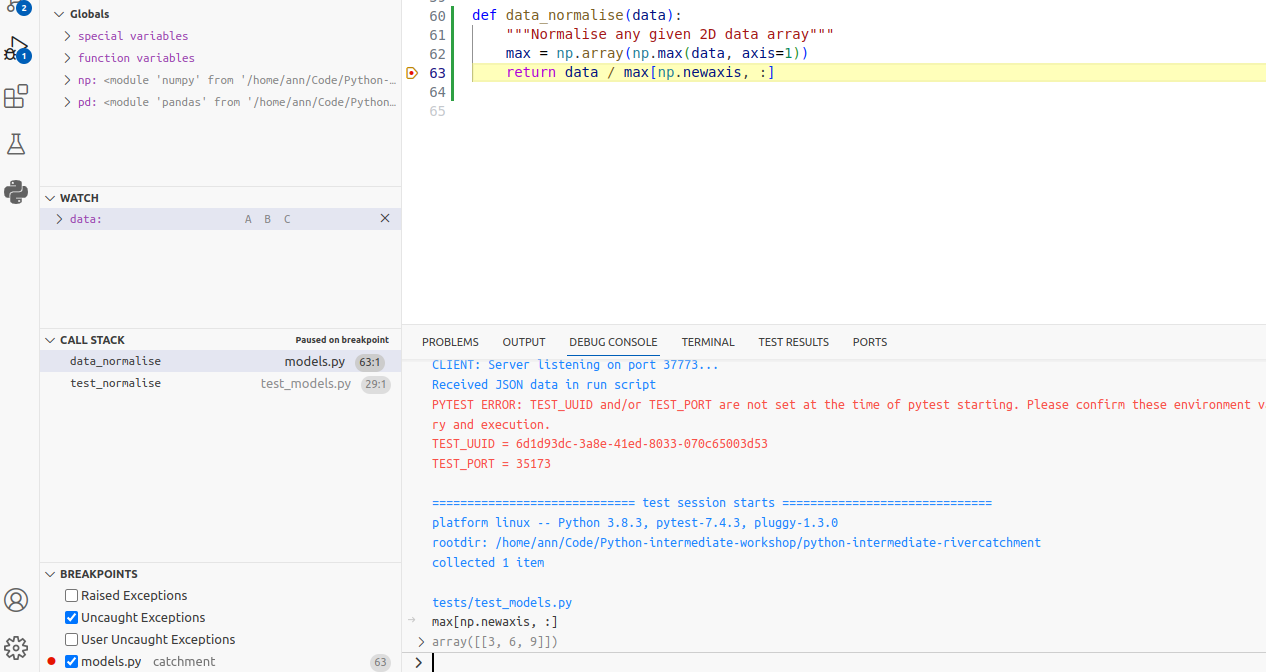
Now, looking at the max variable,
we can see that something looks wrong,
as the maximum values for each patient do not correspond to the data array.
Recall that the input data array we are using for the function is
[[1, 2, 3],
[4, 5, 6],
[7, 8, 9]]
So the maximum value for each measurement set (column) should be [7, 8, 9],
whereas the debugger shows [3, 6, 9].
You can see that the latter corresponds exactly to the last row of data,
and we can immediately conclude that
we took the maximum along the wrong axis of data.
Now we have our answer,
stop the debugging process by selecting
the red square at the top centre of the main VS Code editor window.
So to fix the data_normalise function in models.py,
change axis=1 in the first line of the function to axis=0.
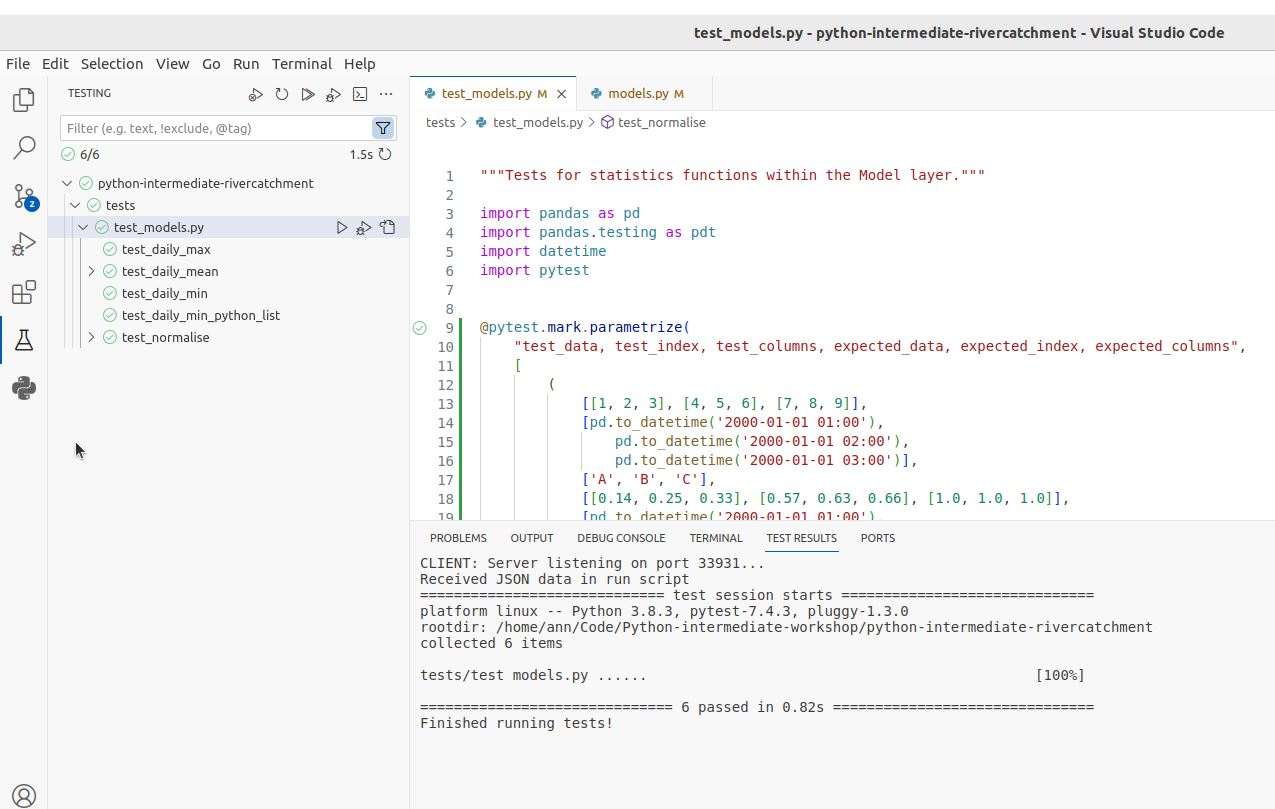
With this fix in place,
running all the tests again should result in all tests passing.
Navigate back to the Testing tab (chemistry flask/beaker icon) on the left hand vertical bar and click on
the arrow next to test_models.py
You should be rewarded with:
NumPy Axis
Getting the axes right in NumPy is not trivial - the following tutorial offers a good explanation on how axes work when applying NumPy functions to arrays.
NumPy vs Pandas: Reducing Test Complexity
So far we have used Pandas testing functions, because the functions we have been testing
make use of Pandas Dataframe functionality. However, even though we will be using the
data_normalise function on Pandas dataframes, the function itself does not actually
require Pandas functionality. The test above demonstrates that the data_normalise
function does not change the input dataframe in any unexpected way; the returned Dataframe
has the same indices and columns as the input Dataframe. Because we know this, we can
simplify the rest of our tests for this function, by using NumPy arrays and testing
functions, instead of the Pandas equivalents. Reducing complexity like this, where you
can, helps you understand what is being tested, and avoid possible confusions.
Before we carry on with new tests, we will reproduce the test above using NumPy, so that you can compare the two testing frameworks. Add an import statement for numpy.testing, and the test test_numpy_normalise, as shown below, to your test_models.py script. Then run the test to confirm it works as expected.
import numpy.testing as npt
...
@pytest.mark.parametrize(
"test, expected",
[
(
[[1, 2, 3], [4, 5, 6], [7, 8, 9]],
[[0.14, 0.25, 0.33], [0.57, 0.63, 0.66], [1.0, 1.0, 1.0]]
)
])
def test_numpy_normalise(test, expected):
"""Test normalisation works for numpy arrays"""
from catchment.models import data_normalise
npt.assert_almost_equal(data_normalise(np.array(test)), np.array(expected), decimal=2)
Note here that we are using the Numpy testing function npt.assert_almost_equal, which allows us to set a relevant test accuracy, using decimal=2. This is equivalent to the atol=1e-2 tolerance setting that we used for the equivalent Pandas test pdt.assert_frame_equal. Numpy also has a testing function npt.assert_array_equal, which tests for exact array matches. The functionality of this test is closely replicated by the default tolerance settings in pd.assert_frame_equal (atol=1e-8 and rtol=1e-5), and can be fully replicated by setting the option check_exact=True when using this function.
Corner or Edge Cases
The test case that we have currently written for data_normalise
is parameterised with a fairly standard data array.
However, when writing your test cases,
it is important to consider parameterising them by unusual or extreme values,
in order to test all the edge or corner cases that your code could be exposed to in practice.
Generally speaking, it is at these extreme cases that you will find your code failing,
so it’s beneficial to test them beforehand.
What is considered an “edge case” for a given component depends on
what that component is meant to do.
In the case of data_normalise function, the goal is to normalise a numeric array of numbers.
For numerical values, extreme cases could be zeros,
very large or small values,
not-a-number (NaN) or infinity values.
Since we are specifically considering an array of values,
an edge case could be that all the numbers of the array are equal.
For all the given edge cases you might come up with,
you should also consider their likelihood of occurrence.
It is often too much effort to exhaustively test a given function against every possible input,
so you should prioritise edge cases that are likely to occur.
For our data_normalise function, some common edge cases might be the occurrence of zeros,
and the case where all the values of the array are the same.
When you are considering edge cases to test for,
try also to think about what might break your code.
For data_normalise we can see that there is a division by
the maximum measurement value for each site,
so this will clearly break if we are dividing by zero here,
resulting in NaN values in the normalised array.
With all this in mind,
let us add a few edge cases to our parametrisation of test_numpy_normalise.
We will add two extra tests,
corresponding to an input array of all 0,
and an input array of all 1.
@pytest.mark.parametrize(
"test, expected",
[
(
[[0.0, 0.0, 0.0], [0.0, 0.0, 0.0], [0.0, 0.0, 0.0]],
[[0.0, 0.0, 0.0], [0.0, 0.0, 0.0], [0.0, 0.0, 0.0]]
),
(
[[1.0, 1.0, 1.0], [1.0, 1.0, 1.0], [1.0, 1.0, 1.0]],
[[1.0, 1.0, 1.0], [1.0, 1.0, 1.0], [1.0, 1.0, 1.0]]
),
(
[[1, 2, 3], [4, 5, 6], [7, 8, 9]],
[[0.14, 0.25, 0.33], [0.57, 0.63, 0.66], [1.0, 1.0, 1.0]]
)
])
def test_numpy_normalise(test, expected):
"""Test normalisation works for numpy arrays"""
from catchment.models import data_normalise
npt.assert_almost_equal(data_normalise(np.array(test)), np.array(expected), decimal=2)
Running the tests now from the command line results in the following assertion error, due to the division by zero as we predicted.
E AssertionError:
E Arrays are not almost equal to 2 decimals
E
E x and y nan location mismatch:
E x: array([[nan, nan, nan],
E [nan, nan, nan],
E [nan, nan, nan]])
E y: array([[0., 0., 0.],
E [0., 0., 0.],
E [0., 0., 0.]])
tests/test_models.py:160: AssertionError
How can we fix this?
Luckily, there is a NumPy function that is useful here,
np.isnan(),
which we can use to replace all the NaN’s with our desired result,
which is 0.
We can also silence the run-time warning using
np.errstate:
...
def data_normalise(data):
"""
Normalise any given 2D data array
NaN values are replaced with a value of 0
"""
max = np.array(np.max(data, axis=0))
with np.errstate(invalid='ignore', divide='ignore'):
normalised = data / max[np.newaxis, :]
normalised[np.isnan(normalised)] = 0.0
return normalised
...
Exercise: Exploring Tests for Edge Cases
Think of some more suitable edge cases to test our
data_normalise()function and add them to the parametrised tests. Remember to build tests for the functionality we want from the function - it does not matter at the moment if some of the tests fail. After you have finished remember to commit your changes.Possible Solution
@pytest.mark.parametrize( "test, expected", [ ( [[0, 0, 0], [0, 0, 0], [0, 0, 0]], [[0, 0, 0], [0, 0, 0], [0, 0, 0]], ), ( [[1, 1, 1], [1, 1, 1], [1, 1, 1]], [[1, 1, 1], [1, 1, 1], [1, 1, 1]], ), ( [[float('nan'), 1, 1], [1, 1, 1], [1, 1, 1]], [[0, 1, 1], [1, 1, 1], [1, 1, 1]], ), ( [[1, 2, 3], [4, 5, float('nan')], [7, 8, 9]], [[0.14, 0.25, 0.33], [0.57, 0.63, 0.0], [1.0, 1.0, 1.0]], ), ( [[-1, 2, 3], [4, 5, 6], [7, 8, 9]], [[0.0, 0.67, 1], [0.67, 0.83, 1], [0.78, 0.89, 1]], ), ( [[1, 2, 3], [4, 5, 6], [7, 8, 9]], [[0.33, 0.67, 1], [0.67, 0.83, 1], [0.78, 0.89, 1]], ) ]) def test_numpy_normalise(test, expected): """Test normalisation works for numpy arrays of one and positive integers.""" from catchment.models import data_normalise npt.assert_almost_equal(data_normalise(np.array(test)), np.array(expected), decimal=2) ...You could also, for example, test and handle the case of a whole row of NaNs.
Defensive Programming
In the previous section, we made a few design choices for our data_normalise function:
- We are implicitly converting any
NaN, - Normalising a constant 0 array of inflammation results in an identical array of 0s,
- We don’t warn the user of any of these situations.
This could have be handled differently. We might decide that we do not want to silently make these changes to the data, but instead to explicitly check that the input data satisfies a given set of assumptions (e.g. no strings) and raise an error if this is not the case. Then we can proceed with the normalisation, confident that our normalisation function will work correctly.
Checking that input to a function is valid via a set of preconditions
is one of the simplest forms of defensive programming
which is used as a way of avoiding potential errors.
Preconditions are checked at the beginning of the function
to make sure that all assumptions are satisfied.
These assumptions are often based on the value of the arguments, like we have already discussed.
However, in a dynamic language like Python
one of the more common preconditions is to check that the arguments of a function
are of the correct type.
Currently there is nothing stopping someone from calling data_normalise with
a string, a dictionary, or another object that is not a pandas.DataFrame or numpy.ndarray.
As an example, let us change the behaviour of the data_normalise() function
to raise an error on negative inflammation values.
Edit the catchment/models.py file,
and add a precondition check to the beginning of the data_normalise() function like so:
...
if np.any(data < 0):
raise ValueError('Measurement values should not be negative')
...
We can then modify our test function in tests/test_models.py
to check that the function raises the correct exception - a ValueError -
when input to the test contains negative values
(i.e. input case [[-1, 2, 3], [4, 5, 6], [7, 8, 9]]).
The ValueError exception
is part of the standard Python library
and is used to indicate that the function received an argument of the right type,
but of an inappropriate value.
@pytest.mark.parametrize(
"test, expected, expect_raises",
[
... # previous test cases here, with None for expect_raises, except for the next one - add ValueError
... # as an expected exception (since it has a negative input value)
(
[[-1, 2, 3], [4, 5, 6], [7, 8, 9]],
[[0, 0.67, 1], [0.67, 0.83, 1], [0.78, 0.89, 1]],
ValueError,
),
(
[[1, 2, 3], [4, 5, 6], [7, 8, 9]],
[[0.33, 0.67, 1], [0.67, 0.83, 1], [0.78, 0.89, 1]],
None,
),
])
def test_normalise(test, expected, expect_raises):
"""Test normalisation works for arrays of one and positive integers."""
from catchment.models import data_normalise
if expect_raises is not None:
with pytest.raises(expect_raises):
npt.assert_almost_equal(data_normalise(np.array(test)), np.array(expected), decimal=2)
else:
npt.assert_almost_equal(data_normalise(np.array(test)), np.array(expected), decimal=2)
Be sure to commit your changes so far and push them to GitHub.
Optional Exercise: Add a Precondition to Check the Correct Type and Shape of Data
Add preconditions to check that data is a
DataFrameorndarrayobject and that it is of the correct shape. Add corresponding tests to check that the function raises the correct exception. You will find the Python functionisinstanceuseful here, as well as the Python exceptionTypeError. Once you are done, commit your new files, and push the new commits to your remote repository on GitHub.Solution
In
inflammation/models.py:... def data_normalise(data): """ Normalise any given 2D data array NaN values are replaced with a value of 0 :param data: 2D array of inflammation data :type data: ndarray """ if not isinstance(data, np.ndarray) or not isinstance(data, pd.DataFrame): raise TypeError('data input should be DataFrame or ndarray') if len(data.shape) != 2: raise ValueError('data array should be 2-dimensional') if np.any(data < 0): raise ValueError('Measurement values should be non-negative') max = np.nanmax(data, axis=0) with np.errstate(invalid='ignore', divide='ignore'): normalised = data / max[np.newaxis, :] normalised[np.isnan(normalised)] = 0 return normalised ...In
test/test_models.py:... @pytest.mark.parametrize( "test, expected, expect_raises", [ ... ( 'hello', None, TypeError, ), ( 3, None, TypeError, ), ( [[1, 2, 3], [4, 5, 6], [7, 8, 9]], [[0.33, 0.67, 1], [0.67, 0.83, 1], [0.78, 0.89, 1]], None, ) ]) def test_data_normalise(test, expected, expect_raises): """Test normalisation works for arrays of one and positive integers.""" from catchment.models import data_normalise if isinstance(test, list): test = np.array(test) if expect_raises is not None: with pytest.raises(expect_raises): npt.assert_almost_equal(data_normalise(test), np.array(expected), decimal=2) else: npt.assert_almost_equal(data_normalise(test), np.array(expected), decimal=2) ...Note the conversion from
listtonp.arrayhas been moved out of the call tonpt.assert_almost_equal()within the test function, and is now only applied to list items (rather than all items). This allows for greater flexibility with our test inputs, since this wouldn’t work in the test case that uses a string.
If you do the challenge, again, be sure to commit your changes and push them to GitHub.
You should not take it too far by trying to code preconditions for every conceivable eventuality.
You should aim to strike a balance between
making sure you secure your function against incorrect use,
and writing an overly complicated and expensive function
that handles cases that are likely never going to occur.
For example, it would be sensible to validate the shape of your measurement data array
when it is actually read from the csv file (in load_csv),
and therefore there is no reason to test this again in data_normalise.
You can also decide against adding explicit preconditions in your code,
and instead state the assumptions and limitations of your code
for users of your code in the docstring
and rely on them to invoke your code correctly.
This approach is useful when explicitly checking the precondition is too costly.
Improving Robustness with Automated Code Style Checks
Let’s re-run Pylint over our project after having added some more code to it. From the project root do:
$ pylint catchment
You may see something like the following in Pylint’s output:
************* Module catchment.models
...
catchment/models.py:60:4: W0622: Redefining built-in 'max' (redefined-builtin)
...
The above output indicates that by using the local variable called max
in the data_normalise function,
we have redefined a built-in Python function called max.
This isn’t a good idea and may have some undesired effects
(e.g. if you redefine a built-in name in a global scope
you may cause yourself some trouble which may be difficult to trace).
Exercise: Fix Code Style Errors
Rename our local variable
maxto something else (e.g. call itmax), then rerun your tests and commit these latest changes and push them to GitHub using our usual feature branch workflow. Make sure yourdevelopandmainbranches are up to date.
It may be hard to remember to run linter tools every now and then.
Luckily, we can now add this Pylint execution to our continuous integration builds
as one of the extra tasks.
Since we’re adding an extra feature to our CI workflow,
let’s start this from a new feature branch from the develop branch:
$ git switch -c pylint-ci develop # note a shorthand for creating a branch from another and switching to it
Then to add Pylint to our CI workflow,
we can add the following step to our steps in .github/workflows/main.yml:
...
- name: Check style with Pylint
run: |
python3 -m pylint --fail-under=0 --reports=y catchment
...
Note we need to add --fail-under=0 otherwise
the builds will fail if we don’t get a ‘perfect’ score of 10!
This seems unlikely, so let’s be more pessimistic.
We’ve also added --reports=y which will give us a more detailed report of the code analysis.
Then we can just add this to our repo and trigger a build:
$ git add .github/workflows/main.yml
$ git commit -m "Add Pylint run to build"
$ git push
Then once complete, under the build(s) reports you should see an entry with the output from Pylint as before, but with an extended breakdown of the infractions by category as well as other metrics for the code, such as the number and line percentages of code, docstrings, comments, and empty lines.
So we specified a score of 0 as a minimum which is very low. If we decide as a team on a suitable minimum score for our codebase, we can specify this instead. There are also ways to specify specific style rules that shouldn’t be broken which will cause Pylint to fail, which could be even more useful if we want to mandate a consistent style.
We can specify overrides to Pylint’s rules in a file called .pylintrc
which Pylint can helpfully generate for us.
In our repository root directory:
$ pylint --generate-rcfile > .pylintrc
Looking at this file, you’ll see it’s already pre-populated.
No behaviour is currently changed from the default by generating this file,
but we can amend it to suit our team’s coding style.
For example, a typical rule to customise - favoured by many projects -
is the one involving line length.
You’ll see it’s set to 100, so let’s set that to a more reasonable 120.
While we’re at it, let’s also set our fail-under in this file:
...
# Specify a score threshold to be exceeded before program exits with error.
fail-under=0
...
# Maximum number of characters on a single line.
max-line-length=120
...
Don’t forget to remove the --fail-under argument to Pytest
in our GitHub Actions configuration file too,
since we don’t need it anymore.
Now when we run Pylint we won’t be penalised for having a reasonable line length. For some further hints and tips on how to approach using Pylint for a project, see this article.
Before moving on, be sure to commit all your changes
and then merge to the develop and main branches in the usual manner,
and push them all to GitHub.
Key Points
Unit testing can show us what does not work, but does not help us locate problems in code.
Use a debugger to help you locate problems in code.
A debugger allows us to pause code execution and examine its state by adding breakpoints to lines in code.
Use preconditions to ensure correct behaviour of code.
Ensure that unit tests check for edge and corner cases too.
Using linting tools to automatically flag suspicious programming language constructs and stylistic errors can help improve code robustness.