RStudio Project and Experimental Data
Last updated on 2026-03-03 | Edit this page
Overview
Questions
- How do you use RStudio project to manage your analysis project?
- What is the most effective way to organize directories for an analysis project?
- How to download a dataset from the internet and save it as a file.
Objectives
- Create an RStudio project and the directories required for storing the files pertinent to an analysis project.
- Download the data set that will be used for the subsequent episodes.
Introduction
Typically, an analysis project begins with dataset files, a handful
of R scripts and output files in a directory. As the project advances,
complexity inevitably rises with the addition of more scripts, output
files, and possibly new datasets. The complexity is further amplified
when dealing with multiple versions of scripts and output files,
necessitating efficient organisation. If these are not well-managed from
the beginning, resuming the project after a break, or sharing the
project with someone else becomes challenging and time-consuming, as we
struggle to recall the project’s status and navigate the directory tree.
Additionally, without proper organisation, the project’s complexity
could lead to frequent use of the setwd function to switch
between different working directories, resulting in a disorganised
workspace.
In this lesson, we will first focus on an effective strategy for managing the files, both used and generated by our data analysis project, within a working directory.
What is a working directory?
A working directory in R is the default location on your computer where R looks for files to load or store any data you wish to save. More information can be found in our Introduction to data analysis with R and Bioconductor lesson.
Secondly, we will also learn how to leverage RStudio projects, a feature built-in to RStudio for managing our analysis project.
What is RStudio?
RStudio is a freely available integrated development environment (IDE), widely used by scientists and software developers for developing software or analysing datasets in R. If you require assistance with RStudio or its general usage, please refer to our Introduction to data analysis with R and Bioconductor lesson.
Finally, in this lesson, we will also learn to use the R function
download.file to download the data for the subsequent
episodes.
Structuring your working directory
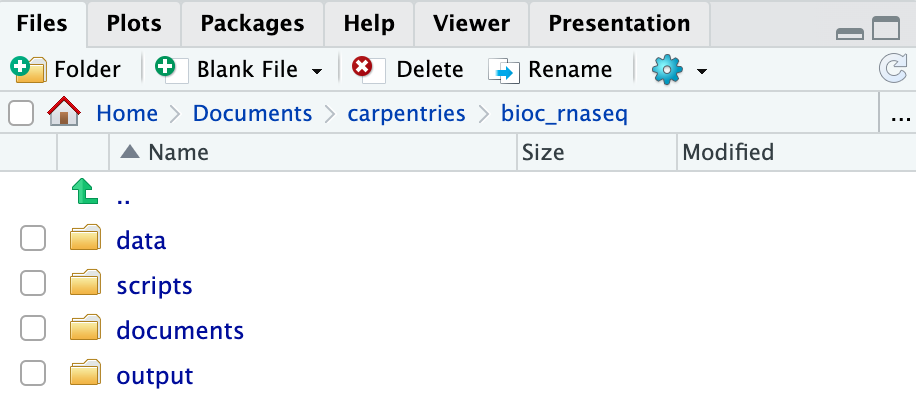
For a more streamlined workflow, we suggest storing all files associated with an analysis in a specific directory, which will serve as your project’s working directory. Initially, this working directory should contain four distinct directories:
-
data: dedicated to storing raw data. This folder should ideally only house raw data and not be modified unless you receive a new dataset (even then, if you have the storage capacity, we suggest you retain the previous dataset as well in case you need it again in the future). For RNA-seq data analysis, this directory will typically contain*.fastqfiles and any related metadata files for the experiment. -
scripts: for storing the R scripts you’ve written and utilised for analysing the data. -
documents: for storing documents related to your analysis, such as a manuscript outline or meeting notes with your team. -
output: for storing intermediate or final results generated by the R scripts in thescriptsdirectory. Importantly, if you carry out data cleaning or pre-processing, the output should ideally be stored in this directory, as these no longer represent raw data.
As your project grows in complexity, you might find it necessary to create more directories or sub-directories. Nevertheless, the aforementioned four directories should serve as the foundation of your working directory.
Create the directories for subsequent episodes
Create a directory on your computer to serve as the working directory
for the rest of this episode and lesson (the workshop example uses a
directory called bio_rnaseq). Then, within this chosen
directory, create the four fundamental directories previously discussed
(data, scripts, documents, and
output).
Using RStudio project to manage your working directory
As previously highlighted, RStudio project is a feature built-into
RStudio for managing your analysis project. It does so by storing
project-specific settings in an .Rproj file stored in your
project’s working directory. Loading these settings up into RStudio by
either opening the .Rproj file directly or through
RStudio’s open project option (from the menu bar, select
File > Open Project...) will automatically
set your working directory in R to the location of the
.Rproj file, essentially your project’s working
directory.
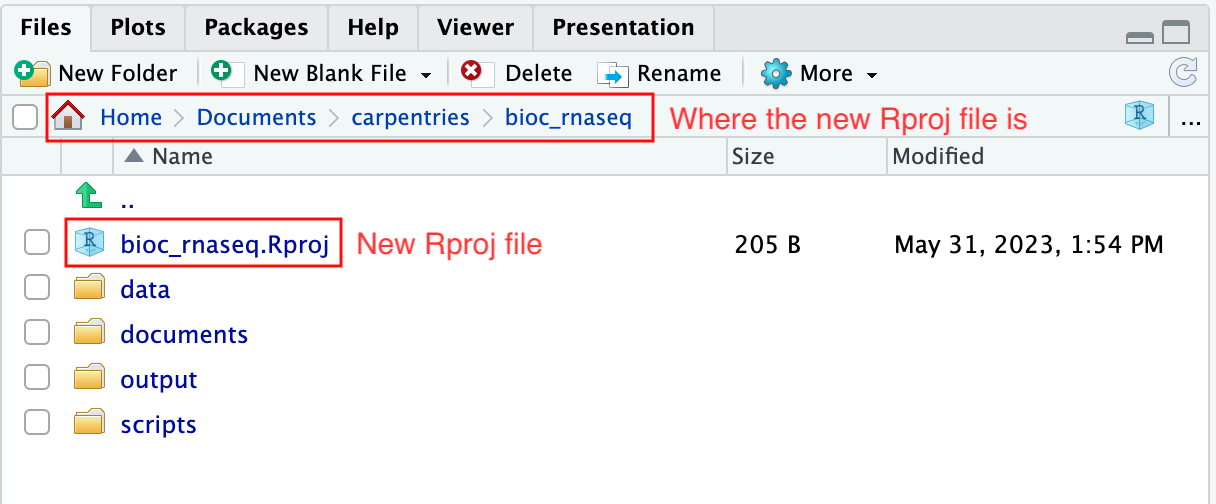
To create an RStudio project:
- Start RStudio.
- Navigate to the menu bar and select
File>New Project.... - Choose
Existing Directory. - Click
Browse...button, and select the directory you have previously chosen as the working directory for the analysis (i.e., the directory where the 4 essential directories reside). - Click
Create Projectat the bottom right of the window.
Upon completion of the steps above, you will find a
.Rproj file within your project’s working directory.
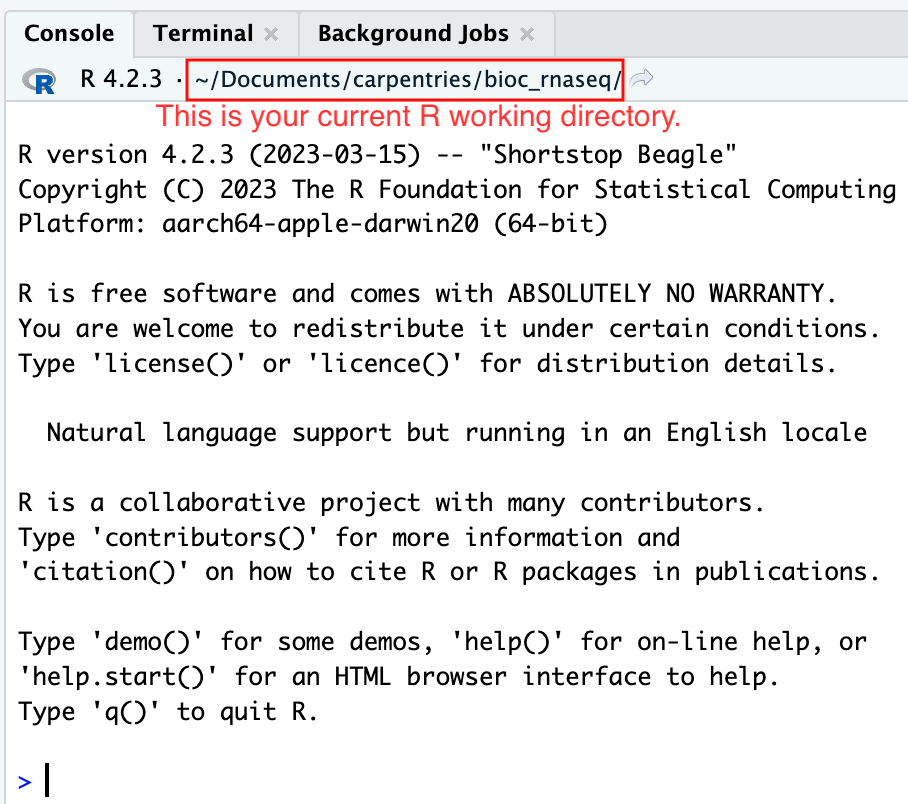
Moreover, the heading of your RStudio console should now also display
the absolute path of your project’s working directory, i.e., where the
.Rproj file resides, indicating that RStudio has set this
directory as your working directory in R.
From this point forward, any R code that you execute that involves reading data from a file or saving data in a file will, by default, be directed to a path relative to your project’s working directory.
If you wish to close the project, perhaps to open another project,
create a new one, or take a break from the project, you can do so by
using to File > Close Project option
located in the menu bar. To open the project back up, either
double-click on the .Rproj file in the working directory, or open up
RStudio and using the File > Open Project
option in the menu bar.
Download the RNA-seq data for subsequent episodes
Finally, we will learn how to use R to download the RNA-seq data required for the subsequent episodes of the lesson. The dataset we will be using was generated to investigate the impact upper-respiratory infection have on changes in RNA transcription in the cerebellum and spinal cord of mice. This dataset was produced as part of the following study:
Blackmore, Stephen, et al. “Influenza infection triggers disease in a genetic model of experimental autoimmune encephalomyelitis.” Proceedings of the National Academy of Sciences 114.30 (2017): E6107-E6116.
The dataset is available at Gene Expression Omnibus (GEO), under the accession number GSE96870. Downloading data from GEO is not straightforward (and won’t be covered in this lesson). Hence, we have made the data available on our GitHub repository for easier access.
To download the files, we will use the R function
download.file, which necessitates at least two parameters:
url and destfile. The url
parameter is used to specify the address on the internet to download the
data from. The destfile parameter indicates where to save
the downloaded file and what the downloaded file should be named.
Let’s download one of the four data files needed for the remainder of
this lesson. The data file is located at https://github.com/carpentries-incubator/bioc-rnaseq/raw/main/episodes/data/GSE96870_counts_cerebellum.csv.
We shall save the downloaded file in the data folder of our
working directory with the name
GSE96870_counts_cerebellum.csv.
R
download.file(
url = "https://github.com/carpentries-incubator/bioc-rnaseq/raw/main/episodes/data/GSE96870_counts_cerebellum.csv",
destfile = "data/GSE96870_counts_cerebellum.csv"
)
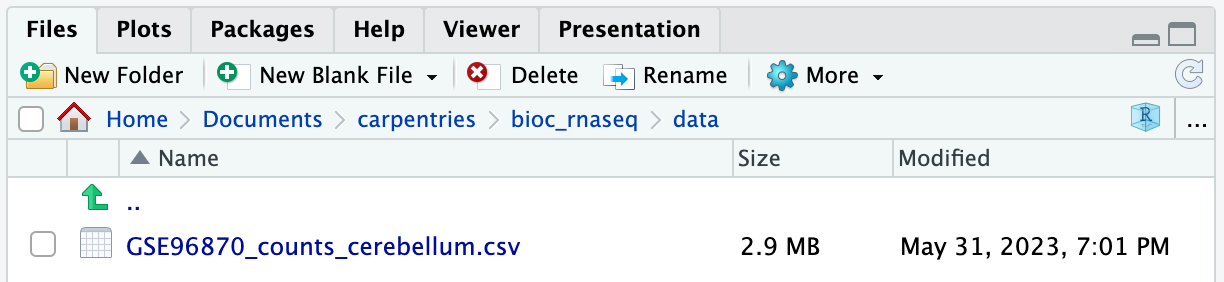
If you navigate to the data folder in your working
directory, you should now find a file named
GSE96870_counts_cerebellum.csv.
Download the remaining data set files
There are three more data set files we need to download for the remainder of this lesson.
| URL | Filename |
|---|---|
| https://github.com/carpentries-incubator/bioc-rnaseq/raw/main/episodes/data/GSE96870_coldata_cerebellum.csv | GSE96870_coldata_cerebellum.csv |
| https://github.com/carpentries-incubator/bioc-rnaseq/raw/main/episodes/data/GSE96870_coldata_all.csv | GSE96870_coldata_all.csv |
| https://github.com/carpentries-incubator/bioc-rnaseq/raw/main/episodes/data/GSE96870_rowranges.tsv | GSE96870_rowranges.tsv |
Use the download.file function to download the files
into the data folder in your working directory.
R
download.file(
url = "https://github.com/carpentries-incubator/bioc-rnaseq/raw/main/episodes/data/GSE96870_coldata_cerebellum.csv",
destfile = "data/GSE96870_coldata_cerebellum.csv"
)
download.file(
url = "https://github.com/carpentries-incubator/bioc-rnaseq/raw/main/episodes/data/GSE96870_coldata_all.csv",
destfile = "data/GSE96870_coldata_all.csv"
)
download.file(
url = "https://github.com/carpentries-incubator/bioc-rnaseq/raw/main/episodes/data/GSE96870_rowranges.tsv",
destfile = "data/GSE96870_rowranges.tsv"
)
- Proper organisation of the files required for your project in a working directory is crucial for maintaining order and ensuring easy access in the future.
- RStudio project serves as a valuable tool for managing your project’s working directory and facilitating analysis.
- The
download.filefunction in R can be used for downloading datasets from the internet.
