Content from What is a reprex and why is it useful?
Last updated on 2026-02-24 | Edit this page
Estimated time: 25 minutes
Welcome to “RRRR, I’m Stuck!” We’re glad you’re here. Let’s first take care of a few setup steps. You should all have followed the setup instructions on the workshop website, and you should have both R and RStudio installed. You should also have the following packages installed: {reprex}, {tidyverse}, and {ggplot2}. If you haven’t installed those packages, please take a moment to install them now. We have a range of levels of experience here. This workshop assumes that you are a researcher in ecology or biology who has some prior experience working with R in RStudio.
This lesson will follow a fictional ecology grad student named Mickey as they explore a new dataset, deal with various common coding problems, and learn how to write a minimal reproducible example to ask for help in community forums. We will not spend much time discussing the code itself, as we will assume you have a basic background.
It’s okay if you’re not an R expert–-but even experienced coders may not always know how to get unstuck, so we hope this workshop will be useful to many levels of coding background.
[MAYBE ADD SOMETHING ABOUT WHAT THE OVERALL LESSON OBKECTIVES ARE]
In this first episode we will introduce the concept of a minimal reproducible example and how it can be useful for getting unstuck while coding. We will provide you with a roadmap that highlights the key steps to follow as we move through the lesson. And we will take a look at the dataset that we’ll be using throughout the rest of the lesson.
Overview
Questions
- What steps can you take to solve problems in your code?
- What is a minimal reproducible example?
- Why are minimal reproducible examples important?
- What is the Portal Project dataset?
Objectives
- Describe a minimal reproducible example and its requirements.
- Recognize how creating a minimal reproducible example can help you solve problems in your code.
- List the key steps to creating a minimal reproducible example.
- Explain the benefits of creating a minimal reproducible example both for you and for others.
- Load in the rodent survey data and briefly explain its contents.
The following exercises are optional, but they can are useful for getting learners settled in. They could happen at the start of the lesson, to break the ice:
“Before we begin, let’s take a moment to reflect on why we are here”
Or they can happen after a first introduction to reprexes.
One of the most frustrating parts of learning to code is getting stuck and not knowing what to do! Maybe R gives you an angry red error message you don’t understand, or your code doesn’t seem to be doing what you were expecting and you don’t know why. Maybe you try to use Google to find answers but you can’t quite find the same problem out there. What to do?
Luckily, there are many people in the R and data science communities who are happy to help. However, in order for them to do so, you must give them the right information. Figuring out how to ask a good question can be hard.
Many helpers or forums may ask you to provide example data or a minimal reproducible example (commonly abbreviated as a “reprex”). What even is that? Wouldn’t it be nice if you could just hand over your computer so a helper can see exactly what is happening? That’s exactly what the reprex is for.
What is a reprex?
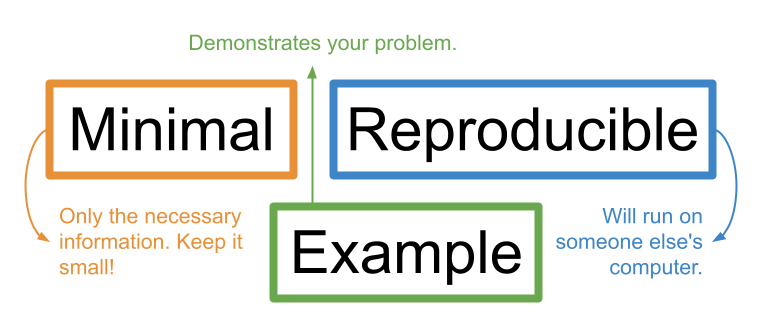
A reprex is essentially a simplified version of your problematic code that clearly demonstrates the problem you are facing (includes only the necessary information to show the problem, nothing more) and will run easily on anyone’s computer.
The Tidyverse documentation puts it simply:
“The goal of a reprex is to package your problematic code in such a way that other people can run it and feel your pain. Then, hopefully, they can provide a solution and put you out of your misery.” - Get help! (Tidyverse)
If you didn’t take time for the reflection exercises at the beginning, you could also pause to do them here.
Why use a reprex?
Reprexes are very important tools to get help when you’re stuck on a coding problem. You may be asked to provide a reprex when you’re working with a statistical consultant (often available at universities) or when posting a question to online help forums (such as StackOverflow or the Posit Community).
As the name suggests, a minimal reproducible example needs to be minimal and reproducible.
Stripping the code and data down to their essential (minimal) parts makes it easy for a helper to zero-in on what might be going wrong.
Making your example reproducible allows a helper to run your code on their own computer so they can easily “tinker” with it to fix it. This makes them more likely to help you.
Helpers
There are lots of people who might help you with your code: friends, colleagues, mentors, or total strangers online. In this lesson, we will use the term “helper” to refer to the person who is helping you to debug your code. Helpers are the target audience for your minimal reproducible example.
But there’s another hidden reason to make a reprex! The process of making a reprex often leads to a better understanding of your own code. Therefore, you might end up solving the problem yourself without asking for help.
Rubber duck debugging
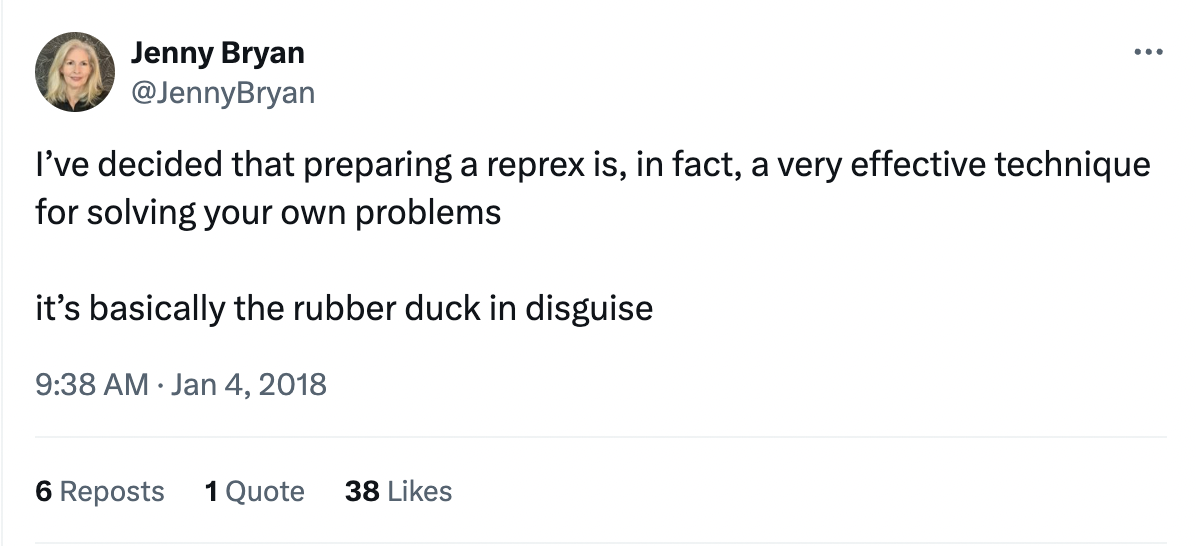
The phenomenon of solving one’s own problem during the process of trying to explain it to someone else is often called “rubber duck debugging.” This is a reference to a story about programmers who would explain the problem they were having with their code to a rubber duck they would keep on their desk. Jenny Bryan refers to reprexes as “basically the rubber duck in disguise,” because they force you to unpack your problem to explain it more clearly.
Jenny Bryan shares many other insights about reprexes in her 2018 talk “Help me help you: Creating reproducible examples.”
Making a reprex can be an excellent learning opportunity, but the process can feel daunting when you are not sure where to begin. In this lesson, we will walk through a step by step roadmap you can use whenever you feel stuck, including some first steps for debugging your code and the process of creating a reprex. We’ll talk about each of the steps and provide a workflow that you can follow when you get stuck in the future. At the end, we’ll introduce you to the {reprex} package, a useful tool for creating good minimal reproducible examples. By the end of the lesson you will have gained a better understanding of how to approach error and warning messages, you will feel more confident in your ability to make a reprex, and you will feel more comfortable asking for formal help.
Meet Mickey, your learning companion
Mickey is an ecology grad student who just joined a new lab. Mickey’s lab has been working for many years with data from the Portal Project, a long-term research study of rodents in Portal, Arizona. Mickey would like to explore this data for their research, so they reach out to Remy, a fifth-year grad student who is very familiar with this project. To get Mickey started, Remy sends Mickey an archival dataset of rodent surveys from 1977-1989, and tells Mickey to “play around” with the data in RStudio to get familiar with it.
Mickey has some past experience in R. They attended the “Data Analysis and Visualization in R for Ecologists” Carpentries workshop, and they feel comfortable with the fundamentals of coding in R. Still, Mickey is a little rusty and nervous about their skills and the unfamiliar data.
Prerequisites and target audience
This workshop assumes some prior experience with working in R and RStudio. We will assume you’ve taken the equivalent of the Data Analysis and Visualization in R for Ecologists workshop and are comfortable with basic commands, and we won’t necessarily explain every line of code in detail.
If you’re much more experienced in R, this workshop is still for you! Even expert coders may not always know how to get unstuck. We hope this workshop will be useful to people with a variety of coding backgrounds.
Mickey starts by loading the data so they can begin to explore it. They also load the {tidyverse}, a set of packages that will be useful for wrangling and visualizing the data.
Loading the entire {tidyverse} here, rather than a few component packages, is an intentional over-complication so that we can teach learners to simplify their packages later. Learners should have {tidyverse} installed, as per the setup instructions. They should also already have a project and folders set up, with the data downloaded and in the “data” folder of the project.
If necessary, here is the url for the data: https://raw.githubusercontent.com/carpentries-incubator/R-help-reprexes/refs/heads/main/episodes/data/surveys_complete_77_89.csv
Let’s go over to RStudio. Make sure that you’re in the RStudio project that you created for this lesson, and that you’ve downloaded the data as a csv and saved it in the “data/” folder.
As a reminder: Make sure you’re coding in your RStudio project. You can open the project you created by double-clicking the “.Rproj” file from your Finder/File Explorer. Or, from inside of RStudio, navigate to the upper right corner of the screen, click on the blue cube icon, choose “Open Project”, and then select your project to open a new session of RStudio.
Now, we can load in the dataset with the following code:
R
# Loading the tidyverse package
library(tidyverse)
R
# Uploading the dataset that is currently saved in the project's data folder
surveys <- read_csv("data/surveys_complete_77_89.csv")
OUTPUT
Rows: 16878 Columns: 13
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (6): species_id, sex, genus, species, taxa, plot_type
dbl (7): record_id, month, day, year, plot_id, hindfoot_length, weight
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Mickey loads in the dataset and takes a look at it to find out what type of data was collected during these surveys.
R
# Take a look at the data
glimpse(surveys)
# or you can use
str(surveys)
OUTPUT
Rows: 16,878
Columns: 13
$ record_id <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,…
$ month <dbl> 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, …
$ day <dbl> 16, 16, 16, 16, 16, 16, 16, 16, 16, 16, 16, 16, 16, 16…
$ year <dbl> 1977, 1977, 1977, 1977, 1977, 1977, 1977, 1977, 1977, …
$ plot_id <dbl> 2, 3, 2, 7, 3, 1, 2, 1, 1, 6, 5, 7, 3, 8, 6, 4, 3, 2, …
$ species_id <chr> "NL", "NL", "DM", "DM", "DM", "PF", "PE", "DM", "DM", …
$ sex <chr> "M", "M", "F", "M", "M", "M", "F", "M", "F", "F", "F",…
$ hindfoot_length <dbl> 32, 33, 37, 36, 35, 14, NA, 37, 34, 20, 53, 38, 35, NA…
$ weight <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ genus <chr> "Neotoma", "Neotoma", "Dipodomys", "Dipodomys", "Dipod…
$ species <chr> "albigula", "albigula", "merriami", "merriami", "merri…
$ taxa <chr> "Rodent", "Rodent", "Rodent", "Rodent", "Rodent", "Rod…
$ plot_type <chr> "Control", "Long-term Krat Exclosure", "Control", "Rod…
spc_tbl_ [16,878 × 13] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
$ record_id : num [1:16878] 1 2 3 4 5 6 7 8 9 10 ...
$ month : num [1:16878] 7 7 7 7 7 7 7 7 7 7 ...
$ day : num [1:16878] 16 16 16 16 16 16 16 16 16 16 ...
$ year : num [1:16878] 1977 1977 1977 1977 1977 ...
$ plot_id : num [1:16878] 2 3 2 7 3 1 2 1 1 6 ...
$ species_id : chr [1:16878] "NL" "NL" "DM" "DM" ...
$ sex : chr [1:16878] "M" "M" "F" "M" ...
$ hindfoot_length: num [1:16878] 32 33 37 36 35 14 NA 37 34 20 ...
$ weight : num [1:16878] NA NA NA NA NA NA NA NA NA NA ...
$ genus : chr [1:16878] "Neotoma" "Neotoma" "Dipodomys" "Dipodomys" ...
$ species : chr [1:16878] "albigula" "albigula" "merriami" "merriami" ...
$ taxa : chr [1:16878] "Rodent" "Rodent" "Rodent" "Rodent" ...
$ plot_type : chr [1:16878] "Control" "Long-term Krat Exclosure" "Control" "Rodent Exclosure" ...
- attr(*, "spec")=
.. cols(
.. record_id = col_double(),
.. month = col_double(),
.. day = col_double(),
.. year = col_double(),
.. plot_id = col_double(),
.. species_id = col_character(),
.. sex = col_character(),
.. hindfoot_length = col_double(),
.. weight = col_double(),
.. genus = col_character(),
.. species = col_character(),
.. taxa = col_character(),
.. plot_type = col_character()
.. )
- attr(*, "problems")=<externalptr> Looking over Mickey’s shoulder, Remy explains that the dataset is
made up of many individual rodent records (record_id). The
date of each record is given by the month,
day, and year columns.
The dataset includes data from a number of different study plots that
had different treatments applied: plot IDs are given by the
plot_id column, and the type of treatment is specified in
plot_type.
There is information about the genus and
species of each individual caught. There is a
species_id column that identifies the species of each
individual caught.
In addition, there is a column called taxa that contains
higher-level taxonomic information. Most of the observations are
rodents, but there are also some birds, rabbits, and reptiles.
R
table(surveys$taxa)
OUTPUT
Bird Rabbit Reptile Rodent
300 69 4 16148 For each individual caught, the field crew took weight,
sex and hindfoot_length measurements when
possible, so values are sometimes missing.
Overall, the dataset contains 16,878 rodent observations ranging across years from 1977 through 1989.
With a clear understanding of the data, Mickey is now free to explore on their own. However, Remy notices that Mickey still looks nervous and decides to share a tool they recently found useful: a roadmap to getting unstuck in R by making a reprex.
Remy’s roadmap outlines four key steps for making a reprex. It is also intended to help the user better understand their problem and potentially find a solution along the way. Remy follows these steps any time they get stuck while coding. Indeed, the first portion of the roadmap, which Remy likes to call “code first aid,” includes preliminary steps to help identify and diagnose the problem, such as determining the type of error, reading function documentation, interpreting error messages, and running through the code line by line.
Remy explains that sometimes, these first aid steps are enough to solve code problems. But if not, the rest of the roadmap will lead Mickey through strategies to better understand the problem and demonstrate it to others in a minimal reproducible example (“reprex”).
Remy emphasizes to Mickey that they are happy to keep helping, but they will be very busy trying to finish writing their dissertation. If Mickey can first follow the steps outlined in this roadmap, then Remy can more easily help with whatever Mickey is struggling to resolve.
With an introduction to the dataset and a roadmap to guide them if they get stuck, Mickey feels ready to start coding!
Mickey feels ready–do your learners also feel ready? This could be a good place to ask what questions they have :)
- Throughout this lesson, we will be walking through a “roadmap” to getting unstuck in R by creating a minimal reproducible example (“reprex”).
- A reprex is a simplified version of your problematic code that clearly demonstrates the problem you are facing and will run on anyone’s computer.
- A reprex should contain only the minimum required to replicate the problem from any device so that helpers can more easily tinker and debug your code.
- The process of building a reprex helps you better understand your code, your data, and your problem so that you will often find the solution yourself!
- The
surveysdataset includes records of rodents captured in a variety of experimental plots over a 12-year period, including some data about each rodent’s sex and morphology.
Content from Identify the problem and make a plan
Last updated on 2026-02-24 | Edit this page
Estimated time: 70 minutes
Overview
Questions
- What do I do when I encounter an error?
- What do I do when my code outputs something I don’t expect?
- Why do errors and warnings appear in R?
- How can I find which areas of code are responsible for errors?
- How can I fix my code? What other options exist if I can’t fix it?
Objectives
After completing this episode, participants should be able to…
- Describe how the desired code output differs from the actual output
- Categorize an error message (e.g. syntax error, semantic errors, package-specific errors, etc.)
- Describe what an error message is trying to communicate
- Identify specific lines and/or functions generating the error message
- Use R Documentation to look up function syntax and examples
- Quickly fix commonly-encountered R errors using ‘code first aid’
- Identify when a problem is better suited for asking for further
help, including making a
reprex
Let’s take a look in more detail at the first step of the roadmap.

In this episode, we’ll cover what to do when you first encounter an error or undesired output from your code. We’ll cover the basics of identifying errors, fixing them if possible, and determining when to create a reprex. At the end of the lesson, we’ll return to Mickey’s analysis.
The first step in solving a problem is understanding what is going wrong. Sometimes R will help you out by displaying an error message when it is unable to run your code. This is a helpful diagnostic tool that, when interpreted correctly, can quickly lead you to a solution. Other times, R doesn’t encounter any problems running your code, but the output is not what you expected. These problems may require a few extra steps to properly diagnose. We will start with easier scenarios and provide helpful “code first aid” steps as we build up to harder challenges.
Strategy 1: Interpret error messages and change function inputs
R will often let us know there’s a problem by displaying an error message. An error that generates an error message is called a syntax error. Error messages happen when R is not able to run your code (this is in contrast to a warning message, which gives a hint that something could be wrong while the code keeps running). Error messages are sometimes straightforward, but other times they can be very tricky to decipher. In this lesson, will teach some tools for interpreting syntax errors for yourself.
Here’s an example error message. In the last episode, we saw that the
taxa column contains information about the higher-level
taxon of each organism caught, such as “Rodent” or “Bird”. We decide to
look at the distribution of taxa by creating a frequency table.
R
table(taxa)
ERROR
Error: object 'taxa' not foundThis code produces an error. What’s going on?
R is telling us that it can’t find taxa in the local
environment. That’s because we haven’t told it where to look–recall that
taxa is the name of a column in the surveys
dataset.
The information from the error message doesn’t specifically tell us
how to solve the error, but it can help us realize what went wrong. In
this case, we can look back at our previous code and see that we were
able to point R to the column using the $ operator:
R
table(surveys$taxa)
OUTPUT
Bird Rabbit Reptile Rodent
300 69 4 16148 That’s better! Now, let’s make a barplot of this column to show this same information.
R
# Make a plot of the different taxa in the rodents dataset
ggplot(aes(x = surveys$taxa)) + geom_bar()
ERROR
Error in `fortify()` at ggplot2/R/plot.R:121:3:
! `data` must be a <data.frame>, or an object coercible by `fortify()`,
or a valid <data.frame>-like object coercible by `as.data.frame()`, not a
<uneval> object.
ℹ Did you accidentally pass `aes()` to the `data` argument?It seems like that should have worked, but we got another error message, and this one seems harder to interpret. Once again, let’s go over each part of the error message and see if it provides any clues.
First, we see an error from a function called fortify,
which we didn’t even use! Then, there’s a more helpful informational
message: “Did you accidentally pass aes() to the
data argument?” This does seem to relate to our line of
code, as we do pass aes into the ggplot
function. But what is this “data argument?”
Strategy 2: Read function documentation
Reading the error message gave us some clues, but it wasn’t enough to fix the problem. Let’s try another strategy. Reading the function documentation can improve our understanding of how the function works, and often that can reveal problems with our code.
Let’s access the documentation for the function
ggplot:
R
?ggplot
A Help window pops up in RStudio. [Add discussion of the different
sections of function documentation, in general terms, here?] The “Usage”
and “Arguments” sections tell us that ggplot takes the argument
data, followed by mapping, which uses
aes(). The “Arguments” section tells us that if the object
passed to that first data argument isn’t already a data
frame, ggplot will try to convert it to a data frame using
fortify. That function, fortify, sounds
familiar from the error message! This gives us an important clue. It
looks like we accidentally passed the mapping argument into
the position where ggplot expected data in the form of a
data frame.
The “Examples” section of the function documentation can be
particularly helpful because it shows how functions are used in context.
The “Examples” section in the ggplot documentation, under
“Pattern 1”, shows exactly how ggplot expects the
data and mapping arguments to be written.
Using this information, we can change our code to put the arguments
in the right order–first the name of the dataset for the
data argument, and then the aes() call for the
mapping argument.
R
ggplot(data = surveys, mapping = aes(x = taxa)) + geom_bar()
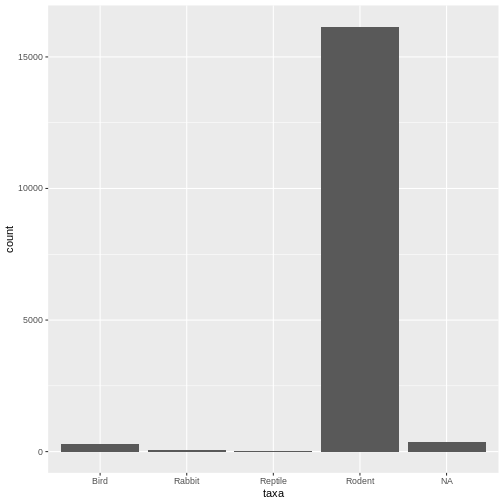
The error message is gone, and it works! Here we see our desired plot. What do you notice about the data?
- Lots of rodents - Some missing values (NAs)
Let’s take a moment to highlight some patterns we’re starting to see in the course of tinkering with our code.
First, we noticed a problem. In this case, the problem was a syntax error, in which the code failed to run and we got an error message.
We carefully read the error message and took a guess at what might be wrong. We changed the inputs to the function accordingly and tried again.
When we encountered another error message, we looked through the function documentation to find more clues about how to fix the error. This helped us see that we were missing an argument.
Another strategy we could have tried would be to copy and paste the error message into a search engine or a generative LLM for more interpretable explanations. And, when all else fails, we can prepare our code into a reproducible example for expert help.
Let’s see if we can use the strategies we’ve learned so far to address a new problem. Here they are, for reference:
Code first aid strategies
- Interpret error messages and tweak inputs
- Look at the function documentation
- Put the error message into a search engine or a generative LLM
Exercise 1: Applying code first aid
Below we see an error message pop up when trying to quantify the
counts of genus and species in our dataset.
Which of the following interpretations of the error message most aptly
describes the problem (hint: look at the ?tally
documentation if you’re stuck!)
R
surveys %>% tally(genus, species)
ERROR
Error in `tally()` at magrittr/R/pipe.R:136:3:
ℹ In argument: `n = sum(genus, na.rm = TRUE)`.
Caused by error in `sum()`:
! invalid 'type' (character) of argumenttallydoes not accept'type' (character)arguments. We should change genus and species to factors or numbers and run this line again.tallydoes not accept'type' (character)arguments. There is no way to quantify these data with this function.tallydoes not accept'type' (character)arguments. We need to assign a weight (e.g. 1) to each row so it knows how much to numerically weigh each observation.tallydoes not accept'type' (character)arguments. This function is not intended togroup_bytwo variables and a different function (count) is required instead.
d is the correct answer!
R
surveys %>% count(genus, species)
OUTPUT
# A tibble: 36 × 3
genus species n
<chr> <chr> <int>
1 Ammospermophilus harrisi 136
2 Amphispiza bilineata 223
3 Baiomys taylori 3
4 Calamospiza melanocorys 13
5 Callipepla squamata 16
6 Campylorhynchus brunneicapillus 23
7 Chaetodipus penicillatus 382
8 Crotalus scutalatus 1
9 Crotalus viridis 1
10 Dipodomys merriami 5675
# ℹ 26 more rowsSemantic errors
Let’s go back to our rodent analysis. We would like to subset the
data to include only the Rodent taxon (as opposed to the
other taxa included in the dataset: Bird, Rabbit, Reptile or NA). Let’s
quickly check to see how much data we’d be throwing out by doing so:
R
table(surveys$taxa)
OUTPUT
Bird Rabbit Reptile Rodent
300 69 4 16148 We’re interested in the rodents, and thankfully it seems like a majority of our observations will be maintained when subsetting to rodents. But wait… In the barplot above, we could clearly see that there were some NA values. Why don’t we see them here?
This is a new type of problem, called a semantic error: the R code ran without any error messages, but it produced an unexpected output. Because there is no error message, semantic errors can be sneaky and hard to notice!
We can’t use our first code first aid strategy here, since there is no error message to read. So let’s jump straight to strategy 2, reading the function documentation.
R
?table
OUTPUT
Help on topic 'table' was found in the following packages:
Package Library
vctrs /home/runner/.local/share/renv/cache/v5/linux-ubuntu-jammy/R-4.5/x86_64-pc-linux-gnu/vctrs/0.6.5/c03fa420630029418f7e6da3667aac4a
base /home/runner/.cache/R/renv/sandbox/linux-ubuntu-jammy/R-4.5/x86_64-pc-linux-gnu/9a444a72
Using the first match ...The documentation for table provides some clues. The
“Usage” and “Arguments” sections show us an argument called
useNA that accepts “no”, “ifany”, and “always”, but it’s
not immediately apparent which one we should use to show our NA values.
When we look at “Examples”, we find something else that looks
helpful:
R
table(a) # does not report NA's
table(a, exclude = NULL) # reports NA's
Aha! So it looks like we can use exclude = NULL to
report NAs in our table. Let’s try that.
R
table(surveys$taxa, exclude = NULL)
OUTPUT
Bird Rabbit Reptile Rodent <NA>
300 69 4 16148 357 Problem solved! Now the NA values show up in the table. We see that by subsetting to the “Rodent” taxa, we would losing about 357 NAs, which themselves could be rodents! However, in this case, it seems a small enough portion to safely omit. Let’s subset our data to the rodent taxon.
R
# Just rodents
rodents <- surveys %>% filter(taxa == "Rodent")
Exercise 2: Syntax vs. semantic errors
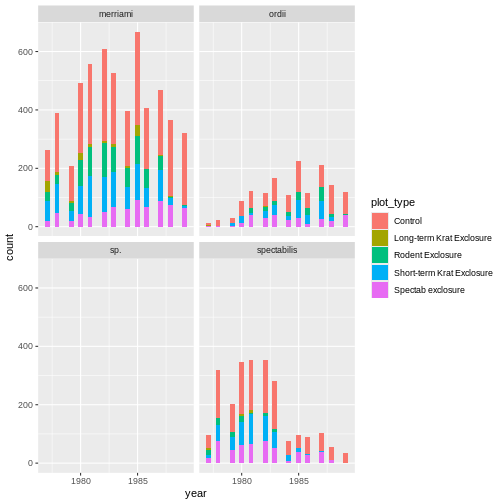
There are 3 lines of code below, and each attempts to create the same plot. Identify which produces a syntax error, which produces a semantic error, and which correctly creates the plot (hint: this may require you inferring what type of graph we’re trying to create!)
ggplot(rodents) + geom_bin_2d(aes(month, plot_type))ggplot(rodents) + geom_tile(aes(month, plot_type), stat = "count")ggplot(rodents) + geom_tile(aes(month, plot_type))
In this case, A correctly creates the graph, plotting as colors in the tile the number of times an observation is seen. It essentially runs the following lines of code:
R
rodents_summary <- rodents %>% group_by(plot_type, month) %>% summarize(count=n())
OUTPUT
`summarise()` has grouped output by 'plot_type'. You can override using the
`.groups` argument.R
ggplot(rodents_summary) + geom_tile(aes(month, plot_type, fill=count))
B is a syntax error, and will produce the following error:
R
ggplot(rodents) + geom_tile(aes(month, plot_type), stat = "count")
ERROR
Error in `geom_tile()`:
! Problem while computing stat.
ℹ Error occurred in the 1st layer.
Caused by error in `setup_params()` at ggplot2/R/ggproto.R:197:17:
! `stat_count()` must only have an x or y aesthetic.Finally, C is a semantic error. It does produce a plot, which is rather meaningless:
R
ggplot(rodents) + geom_tile(aes(month, plot_type))
The steps to identifying the problem and in code first aid matches what we’ve seen above. However, here seeing the problem arise in our code may be much more subtle, and comes from us recognizing output we don’t expect or know to be wrong. Even if the code is run, R may give us warning or informational messages which pop up when executing your code. Most of the time, however, it’s up to the coder to be vigilant and be sure steps are running as they should. Interpreting the problem may also be more difficult as R gives us little or no indication about how it’s misinterpreting our intent.
Generally, the more your code deviates from just using base R
functions, or the more you use specific packages, both the quality of
documentation and online help available from search engines and Googling
gets worse and worse. While base R errors will often be solvable in a
couple of minutes from a quick ?help check or a long online
discussion and solutions on a website like Stack Overflow, errors
arising from little-used packages applied in bespoke analyses might
merit isolating your specific problem to a reproducible example for
online help, or even getting in touch with the developers! Such
community input and questions are often the way packages and
documentation improves over time.
Identifying the problem
In the previous section, it was evident which function was causing a syntax or semantic error. But sometimes, identifying the problem may be trickier. It may be difficult to determine which lines or sections of code are producing the error.
Let’s take a look at another code example. Our goal: to see which k-rat species appear in different plot types over the years.
The misspelling of “Dipodomys” below is deliberate!
R
# Example: identifying a more complex error
# Just k-rats
krats <- rodents %>% filter(genus == "Dipadomys") #filter the dataset to just include the kangaroo rat genus
# plot a histogram of how many observations are seen in each plot type over an x axis of years.
ggplot(krats, aes(year, fill = plot_type)) +
geom_histogram() +
facet_wrap(~species)
ERROR
Error in `combine_vars()` at ggplot2/R/facet-wrap.R:186:5:
! Faceting variables must have at least one value.Uh-oh. Another error here, when we try to make a ggplot. But what is “combine_vars?” And then: “Faceting variables must have at least one value” What does that mean?
This is not an easily interpretable error message from
ggplot, and our code looks like it should run.
This time we put the data argument in the right place, we
included aes(), and we didn’t misspell anything.
Considering this chunk of code, let’s take a step back. Is it
possible that we’re looking at the wrong code? What if the error isn’t
in the ggplot code itself? Let’s look at the krats dataset
to make sure it looks normal.
R
krats
OUTPUT
# A tibble: 0 × 13
# ℹ 13 variables: record_id <dbl>, month <dbl>, day <dbl>, year <dbl>,
# plot_id <dbl>, species_id <chr>, sex <chr>, hindfoot_length <dbl>,
# weight <dbl>, genus <chr>, species <chr>, taxa <chr>, plot_type <chr>It’s empty! Something must have gone wrong not with
ggplot, but with the code we used to create the
krats object.
Strategy 4: Using print() to show information
We can use a print statement to see which genera are
included in the original rodents dataset.
R
print(rodents %>% count(genus))
OUTPUT
# A tibble: 12 × 2
genus n
<chr> <int>
1 Ammospermophilus 136
2 Baiomys 3
3 Chaetodipus 382
4 Dipodomys 9573
5 Neotoma 904
6 Onychomys 1656
7 Perognathus 553
8 Peromyscus 1271
9 Reithrodontomys 1412
10 Rodent 4
11 Sigmodon 103
12 Spermophilus 151This tells us two things. For one, we noticed that we have misspelled Dipodomys, which we can now fix.
Our print() call also tells us that we should expect a
data frame with 9573 values after subsetting to the genus
Dipodomys. This will be useful to check our work after fixing
the misspelling.
R
# Example: identifying a more complex error
# Just k-rats
krats <- rodents %>% filter(genus == "Dipodomys") #filter the dataset to just include the kangaroo rat genus (fixed misspelling)
# check dimensions of krats
dim(krats) # 9573, as expected!
OUTPUT
[1] 9573 13R
# plot a histogram of how many observations are seen in each plot type over an x axis of years.
ggplot(krats, aes(year, fill = plot_type)) +
geom_histogram() +
facet_wrap(~species)
OUTPUT
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.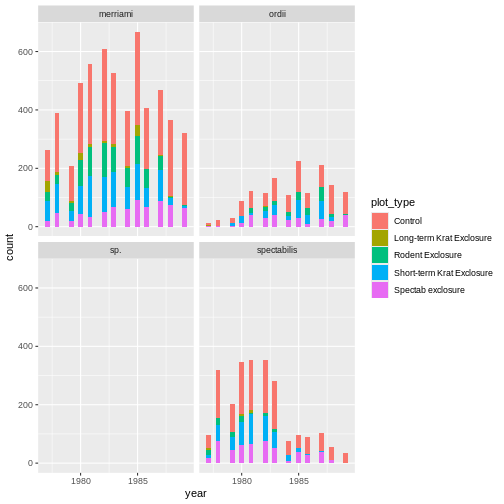
Our improved code here looks good. Checking the dimensions of our
subsetted data frame using dim() function confirms we now
have all the Dipodomys observations, and our plot is looking
better.
Routinely printing out information about your dataset can be a good way to check that your intermediate results make sense.
Summary: Code first aid
Let’s update our list of code first aid strategies:
Code first aid strategies, detailed version
- Identify the problem area
- add print statements immediately upstream or downstream of problem areas
- check the desired output from functions
- see whether any intermediate output can be further isolated and examined separately
- Act on parts of the error we can understand
- interpreting error messages
- changing input to a function
- checking on the control flow of code (e.g. for loops, if/else)
- Reading the R documentation for relevant functions
- reading the documentation’s Description, Usage, Arguments, Details
- testing out code from the Examples section
- Quick online help with a search engine / generative LLM
- Copying error messages for more interpretable explanations
- Describing your error in the hopes of an already-solved solution
- Seeing if an LLM generates equivalent error-free code solving the same goal
We can now understand these steps as a continuous cycle of zeroing in on the problem more and more precisely. Whenever one of the first aid steps helps us identify a part of the code that is failing, we can zoom in on that piece of code and restart the checklist.
If the first aid steps are enough to solve the problem, great! If not, we can stop trying once we get stuck or don’t understand anymore how the code is failing. At that point, we can isolate the specific code area and use it to create a reprex in order to get help from someone else. We’ll delve more into the process of creating a reprex in the next episode.
Exercise 3: Isolating the problem
The following lines of code are not working correctly.
A. What type of error is this? B. Using the toolbox of code first aid strategies, can you isolate the problem area?
R
# Goal: run a chi square test to see whether kangaroo rat observations in the `control` plot type differ significantly between different plot_ids (hopefully not!)
control_plot_data <- krats %>% filter(plot_type == "Control")
n_control_plots <- length(control_plot_data$plot_id)
exp_proportions <- rep(1/n_control_plots, n_control_plots)
plot_counts <- control_plot_data %>% group_by(plot_id) %>% summarize(n = n())
# Chisq test -- do count values vary significantly by plot id?
chisq.test(plot_counts$n, p = exp_proportions)
ERROR
Error in chisq.test(plot_counts$n, p = exp_proportions): 'x' and 'p' must have the same number of elementsAn isolated version of the problem area might look like:
R
n_control_plots <- length(control_plot_data$plot_id)
exp_proportions <- rep(1/n_control_plots, n_control_plots)
# Chisq test -- do count values vary significantly by plot id?
chisq.test(plot_counts$n, p = exp_proportions)
ERROR
Error in chisq.test(plot_counts$n, p = exp_proportions): 'x' and 'p' must have the same number of elementsIf we decide to move the plot_counts line right after the first
control_plot_data line, as plot_counts seems to be calculated correctly.
Here, we can see there’s probably something wrong with the
p argument: exp_proportions is very long, much longer than
the number of control plots! Let’s solve the problem.
R
n_control_plots <- length(unique(control_plot_data$plot_id))
exp_proportions <- rep(1/n_control_plots, n_control_plots)
# Chisq test -- do count values vary significantly by plot id?
chisq.test(plot_counts$n, p = exp_proportions)
OUTPUT
Chi-squared test for given probabilities
data: plot_counts$n
X-squared = 79.977, df = 7, p-value = 1.392e-14We can see that some plots have significantly more or fewer counts than others! Observations of kangaroo rats are not random – rather, some plots seem to attract the kangaroo rats more than others.
When should I prepare my code for a reprex?
If you’ve isolated the problem area and tried using code first aid strategies, but the error persists, it may be time to get some help.
In a classroom setting, we may be used to raising our hand, pointing at our code, and saying “I’m not sure what’s wrong.” But outside of the classroom, helpers have limited time, bandwidth, and requisite knowledge to help. That’s why reproducing the problem with a reproducible example is an essential skill to getting unstuck: it allows you to ask for expert help with a problem that’s clearly identified, self-contained, and reproducible, and allows the expert to quickly see whether they’ve got the requisite skills to answer your question!
Back to our analysis: Mickey tries to get unstuck
Back in the lab, Mickey is happily coding along, exploring the data. Let’s follow their analysis and see how they use code first aid and prepare the code for a reprex.
Mickey is interested in understanding how kangaroo rat weights differ across species and sexes, so they create a quick visualization.
R
# Barplot of rodent species by sex
ggplot(surveys, aes(x = species, fill = sex)) +
geom_bar()
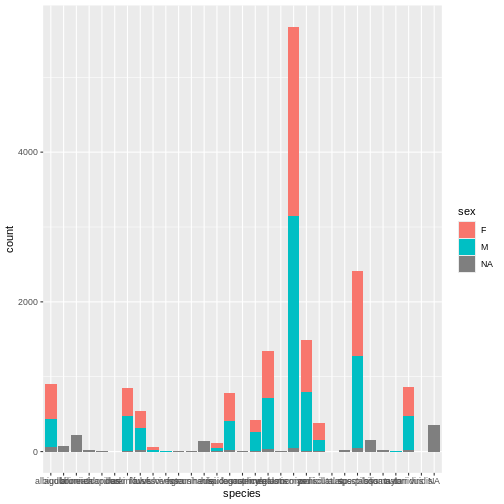
Whoa, this is really overwhelming! Mickey forgot that the dataset includes data for a lot of different species, not just kangaroo rats. Mickey is only interested in two kangaroo rat species: Dipodomys ordii (Ord’s kangaroo rat) and Dipodomys spectabilis (Banner-tailed kangaroo rat).
Mickey also notices that there are three categories for sex: F, M, and what looks like a blank field when there is no sex information available. For the purposes of comparing weights, Mickey wants to focus only rodents of known sex.
Mickey filters the data to include only the two focal species and only rodents whose sex is F or M.
R
# Filter to focal species and known sex
rodents_subset <- surveys %>%
filter(species == c("ordii", "spectabilis"),
sex == c("F", "M"))
Because these scientific names are long, Mickey also decides to add
common names to the dataset. They start by creating a data frame with
the common names, which they will then join to the
rodents_subset dataset:
R
# Add common names
common_names <- data.frame(species = unique(rodents_subset$species), common_name = c("Ord's", "Banner-tailed"))
common_names
OUTPUT
species common_name
1 spectabilis Ord's
2 ordii Banner-tailedBut looking at the common names dataset reveals a
problem! The common names are not properly matched to the scientific
names. For example, the genus Ordii should correspond to Ord’s
kangaroo rat, but currently, it is matched with the Banner-tailed
kangaroo rat instead.
Challenge
- Is this a syntax error or a semantic error? Explain why.
- What “code first aid” steps might be appropriate here? Which ones are unlikely to be helpful?
Mickey re-orders the names and tries the code again. This time, it
works! The common names are joined to the correct scientific names.
Mickey joins the common names to rodents_subset.
R
# Try again, re-ordering the common names
common_names <- data.frame(species = sort(unique(rodents_subset$species)), common_name = c("Ord's", "Banner-Tailed"))
rodents_subset <- left_join(rodents_subset, common_names, by = "species")
Now, Mickey is ready to start learning about kangaroo rat weights.
They start by running a quick linear regression to predict
weight based on species and
sex.
R
# Explore k-rat weights
weight_model <- lm(weight ~ species + sex, data = rodents_subset)
summary(weight_model)
OUTPUT
Call:
lm(formula = weight ~ species + sex, data = rodents_subset)
Residuals:
Min 1Q Median 3Q Max
-109.531 -7.991 3.239 11.469 48.469
Coefficients: (1 not defined because of singularities)
Estimate Std. Error t value Pr(>|t|)
(Intercept) 47.991 1.136 42.23 <2e-16 ***
speciesspectabilis 73.540 1.420 51.79 <2e-16 ***
sexM NA NA NA NA
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 20.58 on 910 degrees of freedom
(31 observations deleted due to missingness)
Multiple R-squared: 0.7466, Adjusted R-squared: 0.7464
F-statistic: 2682 on 1 and 910 DF, p-value: < 2.2e-16The negative coefficient for common_nameOrd's tells
Mickey that Ord’s kangaroo rats are significantly less heavy than
Banner-tailed kangaroo rats.
But something is wrong with the coefficients for sex. Why are there
NA values for sexM? Let’s directly visualize weight by
species and sex to see.
R
# Weight by species and sex
rodents_subset %>%
ggplot(aes(y = weight, x = species, fill = sex)) +
geom_boxplot()
WARNING
Warning: Removed 31 rows containing non-finite outside the scale range
(`stat_boxplot()`).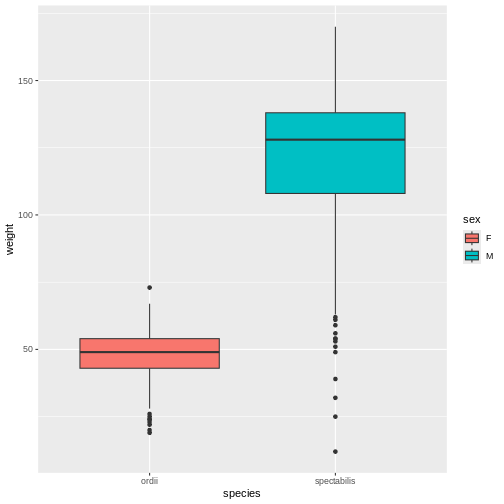
When Mickey visualizes the data, they see a problem in the graph, too. As the model showed, Ord’s kangaroo rats are significantly smaller than Banner-tailed kangaroo rats. But something is definitely wrong! Because the boxes are colored by sex, we can see that all of the Banner-tailed kangaroo rats are male and all of the Ord’s kangaroo rats are female. That can’t be right! What are the chances of catching all one sex for two different species?
To verify that the problem comes from the data, not from the plot
code, Mickey creates a two-way frequency table, which confirms that
there are no observations of female spectabilis or male
ordii in rodents_subset. Something definitely
seems wrong. Those rows should not be missing.
R
# Subsetted dataset
table(rodents_subset$sex, rodents_subset$species)
OUTPUT
ordii spectabilis
F 333 0
M 0 610To double check, Mickey looks at the original dataset.
R
# Original dataset
table(rodents$sex, rodents$species)
OUTPUT
albigula eremicus flavus fulvescens fulviventer harrisi hispidus
F 474 372 222 46 3 0 68
M 368 468 302 16 2 0 42
leucogaster maniculatus megalotis merriami ordii penicillatus sp.
F 373 160 637 2522 690 221 4
M 397 248 680 3108 792 155 5
spectabilis spilosoma taylori torridus
F 1135 1 0 390
M 1232 1 3 441Not only were there originally males and females present from both ordii and spectabilis, but the original numbers were way, way higher! It looks like somewhere along the way, Mickey lost a lot of observations.
While we don’t have the time today, let’s assume Mickey worked their way through the code first aid steps, but weren’t able to solve the problem.
They decide to return to Remy’s road map to figure out what to do next.
Since code first aid was not enough to solve this problem, it looks like it’s time to ask for help using a reprex.
- The first step to getting unstuck is identifying a problem, isolating the problem area, and interpreting the problem
- Often, using “code first aid” – acting on error messages, looking at data, inputs, etc., pulling up documentation, asking a search engine or LLM, can help us to quickly fix the error on our own.
- If code first aid doesn’t work, we can ask for help and prepare a reproducible example (reprex) with a defined problem and isolated code
- We’ll cover future steps to prepare a reproducible example (reprex) in future episodes.
Content from Minimal reproducible code
Last updated on 2026-02-24 | Edit this page
Estimated time: 75 minutes
Overview
Questions
- Why is it important to make a minimal code example?
- Which part of my code is causing the problem?
- Which parts of my code should I include in a minimal example?
- How can I tell whether a code snippet is reproducible or not?
- How can I make my code reproducible?
Objectives
- Explain the value of a minimal code snippet.
- Identify packages or other dependencies needed to run the code.
- Simplify a script down to a minimal code example.
- Evaluate whether a piece of code is reproducible as is or not. If not, identify what is missing.
- Edit a piece of code to make it reproducible
When we left off in the previous episode, Mickey had discovered a problem with their code–many kangaroo rat observations were missing from a subset of the data after they filtered the dataset down to the kangaroo rat species of interest.
Mickey tried some code first aid steps but wasn’t able to solve the problem. They consulted Remy’s road map and saw that the next step is to make a reprex.
Making a reprex

Step 1: Minimize the code
Mickey has written a lot of code so far. The code is also a little messy–for example, after fixing the previous errors, they sometimes commented out the old code and kept it for future reference.
Let’s take a look at the script as it stands so far.
Share this file with learners and/or have them copy/paste it exactly into their RStudio sessions–do not try to re-type all this code line by line! There definitely will not be time.
R
# Minimal reproducible example script
# Loading the tidyverse package
library(tidyverse)
# Uploading the dataset that is currently saved in the project's data folder
surveys <- read_csv("data/surveys_complete_77_89.csv")
# Take a look at the data
glimpse(surveys)
# or you can use
str(surveys)
table(surveys$taxa)
# Barplot of rodent species by sex
ggplot(rodents, aes(x = species, fill = sex)) +
geom_bar()
# Filter to focal species and known sex
rodents_subset <- surveys %>%
filter(species == c("ordii", "spectabilis"),
sex == c("F", "M"))
# Add common names
# common_names <- data.frame(species = unique(rodents_subset$species), common_name = c("Ord's", "Banner-tailed"))
# common_names
# Try again, re-ordering the common names
common_names <- data.frame(species = sort(unique(rodents_subset$species)), common_name = c("Ord's", "Banner-Tailed"))
rodents_subset <- left_join(rodents_subset, common_names, by = "species")
# Explore k-rat weights
weight_model <- lm(weight ~ species + sex, data = rodents_subset)
summary(weight_model)
# Weight by species and sex
rodents_subset %>%
ggplot(aes(y = weight, x = species, fill = sex)) +
geom_boxplot()
# Subsetted dataset
table(rodents_subset$sex, rodents_subset$species)
# Original dataset
table(surveys$sex, surveys$species)
Exercise 1: Reflection
As you look at this script and think through trying to debug it, how do you feel?
Mickey’s first instinct is to send the script to Remy and tell them about the error. Imagine that you are Remy, an advanced graduate student whose priority is finishing your dissertation. Your new labmate Mickey has just sent you this script, asking for help debugging it. How do you feel when you get Mickey’s email? What advice might you give Mickey?
When asking someone else for help, it is important to simplify your code as much as possible to make it easier for the helper to understand what is wrong. Simplifying code helps to reduce frustration and overwhelm when debugging an error in a complicated script. The more that we can make the process of helping easy and painless for the helper, the more likely it is that they will take the time to help.
Create a new script
Why do you think it’s a good idea to create a new script?
To make the task of simplifying the code less overwhelming, let’s create a separate script for our reprex. This will let us experiment with simplifying our code while keeping the original script intact.
Let’s create and save a new, blank R script and give it a name, such as “reprex-script.R”
Making an R script
There are several ways to make an R script:
- File > New File > R Script
- Click the white square with a green plus sign at the top left corner of your RStudio window
- Use a keyboard shortcut: Cmd + Shift + N (on a Mac) or Ctrl + Shift + N (on Windows)
Let’s go ahead and copy over all of our code so we have an exact copy of the full analysis script. This way, we can make as many changes to it as we want and still keep the original code untouched.
Now, we will follow an iterative process to simplify our script.
A. Identify the symptom of the problem. What are you observing that shows you something is wrong?
B. Remove some code that is not central to demonstrating the problem.
C. Run the simplified code and make sure that the symptom is still present. Does your example still reproduce the problem?
A. Identify a symptom of the problem
Let’s figure out which line of code, when you run it, clearly shows that something is wrong. For a syntax error, this is straightforward: it’s the line of code that generates the error message. But our error here is a semantic error. The code runs, but it returns the wrong result. So let’s think instead about what line of code created a result that we could clearly see was incorrect.
This is a little tricky in our case, because we first noticed something was wrong when we looked at the output of the linear model. That model output could be a perfectly reasonable symptom to use!
R
summary(weight_model)
ERROR
Error: object 'weight_model' not foundBut let’s not discount the work we’ve already done to diagnose this problem! Something looked strange about this model, so we made a plot. Something looked strange about the plot, so we double checked the dataset used to create both the model and the plot. By comparing that dataset with the original, un-subsetted data, we were able to determine that something was wrong.
To summarize, we have already determined:
The problem: There are many observations missing
from rodents_subset that should not have been removed.
The symptom (lines that show that something is wrong): Comparison between the species and sex counts in the original and subsetted datasets.
In particular, this comparison shows us that there are no
observations of female spectabilis or male ordii in
rodents_subset, but there were plenty in the
original dataset, and that in general, there were many fewer rows for
both species in the subset than the original.
R
# Subsetted dataset
table(rodents_subset$sex, rodents_subset$species)
ERROR
Error: object 'rodents_subset' not foundR
# Original dataset
table(surveys$sex, surveys$species) # there are no observations of female spectabilis or male ordii in `rodents_subset`, even though there were in the original dataset.
OUTPUT
albigula audubonii bilineata brunneicapillus chlorurus clarki eremicus
F 474 0 0 0 0 0 372
M 368 0 0 0 0 0 468
flavus fulvescens fulviventer fuscus gramineus harrisi hispidus leucogaster
F 222 46 3 0 0 0 68 373
M 302 16 2 0 0 0 42 397
leucophrys maniculatus megalotis melanocorys merriami ordii penicillatus
F 0 160 637 0 2522 690 221
M 0 248 680 0 3108 792 155
scutalatus sp. spectabilis spilosoma squamata taylori torridus viridis
F 0 4 1135 1 0 0 390 0
M 0 5 1232 1 0 3 441 0These two lines of code, and the observation we made about them, will be our guide as we simplify the script further.
We can now start removing pieces of code that we believe are not central to our problem. After each removal, we can re-run the code and make sure that our symptom persists. If the symptom changes, we have either solved our problem (yay for rubber duck debugging!) or we removed a line of code that was actually essential to reproducing our problem.
B. Remove some code that is not central to demonstrating the problem.
Let’s start identifying pieces of code to remove. In general, we can
remove code that does not create variables for later use (for example,
exploratory plots, models, or descriptive functions such as
head() or summary()). We can also get rid of
code that adds complexity to the analysis that is not relevant to the
problem at hand.
Let’s start by removing the broken code that we commented out earlier, back when we tried to join the common names and it didn’t work because they were in the wrong order.
Code to remove:
R
# Add common names
# common_names <- data.frame(species = unique(rodents_subset$species), common_name = c("Ord's", "Banner-tailed"))
# common_names
Actually, now that we think about it, those common names are not directly related to the problem at all! The “common_name” column might be useful later on, but for our reprex we can probably remove that part of the code without changing the outcome.
Code to remove:
R
# Try again, re-ordering the common names
common_names <- data.frame(species = sort(unique(rodents_subset$species)), common_name = c("Ord's", "Banner-Tailed"))
rodents_subset <- left_join(rodents_subset, common_names, by = "species")
After removing both of those pieces of code, our script is a little shorter:
R
# Minimal reproducible example script
# Loading the tidyverse package
library(tidyverse)
# Uploading the dataset that is currently saved in the project's data folder
surveys <- read_csv("data/surveys_complete_77_89.csv")
# Take a look at the data
glimpse(surveys)
# or you can use
str(surveys)
table(surveys$taxa)
# Barplot of rodent species by sex
ggplot(surveys, aes(x = species, fill = sex)) +
geom_bar()
# Filter to focal species and known sex
rodents_subset <- surveys %>%
filter(species == c("ordii", "spectabilis"),
sex == c("F", "M"))
# Explore k-rat weights
weight_model <- lm(weight ~ species + sex, data = rodents_subset)
summary(weight_model)
# Weight by species and sex
rodents_subset %>%
ggplot(aes(y = weight, x = species, fill = sex)) +
geom_boxplot()
# Subsetted dataset
table(rodents_subset$sex, rodents_subset$species)
# Original dataset
table(surveys$sex, surveys$species) # there are no observations of female spectabilis or male ordii in `rodents_subset`, even though there were in the original dataset.
C. Run the simplified code and make sure that the symptom is still present. Does your example still reproduce the problem?
Now it’s time to re-run the script to make sure we haven’t removed anything essential. Remember to pay attention to the symptom of the problem at the end and make sure that the essential observation hasn’t changed. Sure enough, those observations are still missing. We have succeeded in simplifying our code while still demonstrating the problem!
Great progress, but this script is still pretty long and complicated. Can we remove more things?
Exercise 2: Minimizing code
Minimizing code is an iterative process. Repeat steps B and C above several more times. Which other lines of code can you remove to make this script more minimal? After removing each part, be sure to re-run the code to make sure that it still reproduces the error.
- The barplot of species and sex (ggplot) can be removed because it generates a plot but does not create any variables that are used later.
- Similarly, our end visualization of weight by species and sex (boxplot) can be removed.
- The weight model and the summary can be removed
- Any other informational functions that could have been run in the
console, such as
table()orprint(),head(), orstr()can be removed. - The essential parts to keep are the lines that access the dataset in
the first place, subset it down to rodents_subset, and then diagnose the
problem (the
table()calls at the end).
After repeating steps B and C over and over again, we arrive at a much more minimal script.
R
# Loading the tidyverse package
library(tidyverse)
surveys <- read_csv("data/surveys_complete_77_89.csv")
# Filter to focal species and known sex
rodents_subset <- surveys %>%
filter(species == c("ordii", "spectabilis"),
sex == c("F", "M"))
# Subsetted dataset
table(rodents_subset$sex, rodents_subset$species)
# Original dataset
table(surveys$sex, surveys$species) # there are no observations of female spectabilis or male ordii in `rodents_subset`, even though there were in the original dataset.
Mickey is really getting the hang of this! They scrutinize the
example to see if there’s anything else that can be cut. They realize
that the code still runs perfectly fine if they remove
library(tidyverse)–since they already loaded the
{tidyverse} package, there should be no need to load it again!
Mickey realizes that they might be able to narrow the example down eeeeeeven more. They try removing the species filter and only filtering by sex. Now the minimal example looks like this:
R
surveys <- read_csv("data/surveys_complete_77_89.csv")
# Filter to known sex
rodents_subset <- surveys %>%
filter(sex == c("F", "M"))
# Subsetted dataset
table(rodents_subset$sex, rodents_subset$species)
# Original dataset
table(surveys$sex, surveys$species) # still missing a lot of rows!
Something is different–we no longer have zero rows for two of the
species/sex combos. But this example still demonstrates our problem.
Remember, we previously stated the problem as “There are many
observations missing from rodents_subset that should not
have been removed.” And sure enough, if we look closely here, we can see
that our species/sex counts have changed from 690 F ordii/792 M ordii
and 1135 F spectabilis/1232 M spectabilis to 333/393 and 568/610,
respectively. The problem persists! We are still mysteriously missing
rows.
If you had chosen to remove the sex filter instead of removing the species filter, the same point would be made. The numbers would be different, but we would still see fewer rows in the subsetted data frame. Either one works!
If you hadn’t noticed that you could simplify this example even further, that would still be okay! Minimizing code is an art, not an exact science. The more minimal you can make your code, the better, but a helper will still have a much easier time working on your problem if you’ve removed some extraneous steps, even if you haven’t narrowed it down 100%. Don’t let the perfect be the enemy of the good!
Okay, so our minimal snippet looks like this:
R
surveys <- read_csv("data/surveys_complete_77_89.csv")
# Filter to known sex
rodents_subset <- surveys %>%
filter(sex == c("F", "M"))
# Subsetted dataset
table(rodents_subset$sex, rodents_subset$species)
# Original dataset
table(surveys$sex, surveys$species) # still missing a lot of rows!
This is great progress! Remy will find this minimal code snippet much more approachable than the long script that Mickey started with.
Exercise 3: Have we made a reprex?
Mickey is really proud of their efforts to minimize the code! They email the minimal code snippet to Remy to ask for help. Remy notices immediately that this code is much easier to read and understand. They open up the script and try to run it in R.
What do you think will happen when Remy tries to run the code from this reprex script?
What should Mickey do next to improve the minimal reproducible example?
We haven’t yet included enough code to allow a helper, such as Remy, to run the code on their own computer. If Remy tries to run the reprex script in its current state, they will encounter errors because they don’t have access to the same R environment that Mickey does.
That’s why it’s so important to include dependencies in your reprex.
Include dependencies
A dependency is a piece of code that other pieces of code depend on in order to function properly.
R code consists primarily of functions and variables. In order to make our minimal examples truly reproducible, we have to give our helpers access to all the functions and all the variables that are necessary to run our code.
First, let’s talk about functions. Functions in R typically come from packages. You can access them by loading the package into your environment.
To make sure that your helper has access to the packages necessary to
run your reprex, you will need to include calls to
library() for whichever packages are used in the code. For
example, if your code uses the function lmer from the
lme4 package, you would have to include
library(lme4) at the top of your reprex script to make sure
your helper has the lme4 package loaded and can run your
code.
Default packages
Some packages, such as {base} and {stats},
are loaded in R by default, so you might not have realized that a lot of
commonly-used functions, such as dim, colSums,
mean, and length actually come from those
packages!
You can see a complete list of the functions that come from the
{base} and {stats} packages by running
library(help = "base") or
library(help = "stats") in your console.
But, you actually don’t need to worry too much about this because
your helpers’ RStudio versions will also have {base} and
{stats} preinstalled!
Let’s do this for our own reprex. We can start by identifying all the functions used, and then we can figure out where each function comes from to make sure that we tell our helper to load the right packages.
Exercise 4: Which packages should we load?
The functions used in our minimal example are
read_csv(), filter(), c(), and
table().
Identify the package that each of the functions comes from and modify
the minimal example so that it explicitly loads those packages.
:::solution library(dplyr) library(readr)
filter() comes from dplyr, and
read_csv()comes from{readr}.c()andtable()come from{base}`,
which is loaded by default, so we don’t need to include a library() call
for this.
Bonus if you notice that we also use the %>%
operator, which comes from dplyr too, so we definitely
need to make sure that dplyr is loaded!
Extra challenge: did we use any other operators? Where do they come from?
:::
We can update our minimal code to include those
library() calls.
R
library(readr)
library(dplyr)
surveys <- read_csv("data/surveys_complete_77_89.csv")
# Filter to known sex
rodents_subset <- surveys %>%
filter(sex == c("F", "M"))
# Subsetted dataset
table(rodents_subset$sex, rodents_subset$species)
# Original dataset
table(surveys$sex, surveys$species) # still missing a lot of rows!
Installing vs. loading packages
We included calls to library() to load the packages we
need. But what if our helper doesn’t have all of these packages
installed? Won’t the code not be reproducible?
Packages need to be installed one time before they can be loaded with
library(). Typically, we don’t include
install.packages() in our code for each of the packages
that we include in the library() calls, because
install.packages() doesn’t need to be repeated every time
the script is run. We can assume that our helper will see
library(specialpackage) and know that they need to go
install “specialpackage” on their own.
Technically, this does make that part of the code not reproducible!
But it’s also more “polite” than explicitly including
install.packages(). Our helper might have their own way of
managing package versions, and forcing them to install a package when
they run our reprex risks messing up their workflow. It is a common
convention to stick with library() and let the helper
figure it out from there.
Exercise 5: Which packages are essential?
In each of the following code snippets, identify the necessary packages (or other code) to make the example reproducible.
weight_model <- lm(weight ~ common_name + sex, data = rodents_subset)
tab_mod(weight_model)mod <- lmer(weight ~ hindfoot_length + (1|plot_type), data = rodents)
summary(mod)rodents_processed <- process_rodents_data(rodents)
glimpse(rodents_processed)This exercise should take about 10 minutes. :::solution a.
lm is part of base R, so there’s no package needed for
that. tab_mod comes from the package sjPlot.
You could add libary(sjPlot) to this code to make it
reproducible. b. lmer is a linear mixed modeling function
that comes from the package lme4. summary is
from base R. You could add library(lme4) to this code to
make it reproducible. c. process_rodents_data is not from
any package that we know of, so it was probably an originally-created
function. In order to make this example reproducible, you would have to
include the definition of process_rodents_data.
glimpse is probably from dplyr, but it’s worth
noting that there is also a glimpse function in the
pillar package, so this might be ambiguous. This is another
reason it’s important to specify your packages–if you leave your helper
guessing, they might load the wrong package and misunderstand your
error!
:::::::::::::::::::::::::::::::::::::::::::
Including library() calls will definitely help Remy run
the code. But this code still won’t work as written because Remy does
not have access to the same objects that Mickey used in the
code. Along with functions, objects are the second type of dependency we
need to watch out for when writing reprexes.
The code as written relies on rodents_subset, which Remy
will not have access to if they try to run the code. That means that
we’ve succeeded in making our example minimal, but it is not
reproducible: it does not allow someone else to reproduce the
problem!
In the next episode, we will learn how to handle perhaps the most thorny part of creating reprexes: dealing with datasets.
Exercise 6: Reflection
Let’s take a moment to reflect on this process.
What’s one thing you learned in this episode? An insight; a new skill; a process?
What is one thing you’re still confused about? What questions do you have?
This exercise should take about 5 minutes.
- Making a reprex is the next step after trying code first aid.
- In order to make a good reprex, it is important to simplify your code
- Simplify code by removing parts not directly related to the question
- Give helpers access to the functions used in your code by loading all necessary packages
Content from Minimal reproducible data
Last updated on 2026-02-24 | Edit this page
Estimated time: 94 minutes
Overview
Questions
- What is a minimal reproducible dataset, and why do I need it?
- How do I create a minimal reproducible dataset?
- Can I just use my own data?
Objectives
- Describe a minimal reproducible dataset
- Appreciate why it is important to provide example data that is minimal and reproducible
- Recognize that there are different approaches to providing example data
- Identify the relevant aspects of your data
- Create a suitable reprex dataset from scratch
- Share your original dataset in a way that is minimal and reproducible
- Subset a built-in dataset to use in your reprex
4.1 What is a minimal reproducible dataset and why do I need it?
Development note: Previous episodes should have already introduced the concept of a minimal reproducible examples, why it is important, and talked about making the code minimal and reproducible. This data episode is part of being reproducible and should perhaps be followed-up with more details on reproducibility if not previously mentioned (e.g., reprex needs minimal code - check - dependencies, which include minimal data - check - and other basic information like your system and R version as well as contextual information on your data?).
Mickey has now (1) narrowed down their problem area (they are losing observations during filtering), (2) stripped down their code to make it minimal, and have been working on making it reproducible by including all necessary dependencies (so anyone can simply copy-paste the code into their system to replicate their problem). They have done a good job so far, but there is still one dependency missing, can you guess what it is?
Exercise 1
If Remy received the current script and tried to run it as-is, would it work? If not, why?
Below is a reminder of what Mickey’s script currently looks like.
R
# Mickey's current minimal code
library(readr)
library(dplyr)
surveys <- read_csv("data/surveys_complete_77_89.csv")
OUTPUT
Rows: 16878 Columns: 13
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (6): species_id, sex, genus, species, taxa, plot_type
dbl (7): record_id, month, day, year, plot_id, hindfoot_length, weight
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.R
# Filter to known sex
rodents_subset <- surveys %>%
filter(sex == c("F", "M"))
# Subsetted dataset
table(rodents_subset$sex)
OUTPUT
F M
3656 4145 R
# Original dataset
table(surveys$sex) # the numbers don't match!
OUTPUT
F M
7318 8260 The code would fail to run because Remy probably doesn’t have the “surveys_complete_77_89.csv” file, or, more specifically doesn’t have that file in the specified “data” folder. Indeed, we have no idea what Remy’s file management system looks like!
A reprex code will always require a data object in order to run!
To make sure Remy can work on the reprex anywhere, Mickey needs to ensure Remy has the required dataset to run it.
As with the code, a reprex’s dataset should also be minimal–free of unnecessary information and reproducible–such that anyone can recreate it.
Indeed, Step 3 on the roadmap tells us to create a minimal reproducible dataset. However, Mickey figures it would be faster and easier to send Remy the current minimal code along with the original csv file. After all, the code already includes how to read-in the csv file, right? There is no need to create another dataset.
Exercise 2: Reflect
Mickey feels like sharing the original data file would be easier, but who would find it easier, Mickey or Remy?
Mickey is thinking that it would be easier for themselves, not necessarily for Remy.
Remember: one of the goals of creating a reprex is to help the helpers. They don’t have to help, they are volunteering their time. As such, they deserve to be treated with kindness and respect. If you send a code with a separate file, not only is the code not reproducible, but you are creating extra work for the helper. Extra unnecessary and potentially time-consuming steps are more likely to make helpers frustrated rather than happy to help.
If you find yourself getting frustrated at how much time and effort creating a reprex might be taking, remember that (1) trusting the process may reveal the solution along the way; and (2) being kind, clear, and helpful will reward you with a quicker, more accurate solution (and will make it more likely that a helper will help again in the future).
Remy’s feelings aside, while this strategy could still work in this particular instance, there are many reasons why sharing original data may not be possible or recommended. Can you think of any? See callout below for examples.
Think twice before sharing your data!
Even though there are times when sharing your original dataset seems like the easiest approach, there are several reasons why sharing original data may not be possible or recommended.
The original dataset may be:
- too large - the Portal dataset is ~35,000 rows with 13 columns and contains data for decades. That’s a lot!
- private - the dataset might not be published yet, it may not be yours to share, or maybe it includes protected information such as personal medical information or the location of endangered species.
- hard to send - on most online forums, you can’t attach supplemental files (more on this later). Even if you are just sending data to a colleague, file paths can get complicated, the data might be too large to attach, etc.
- complicated - it would be hard to locate the relevant information.
One example to steer away from are needing a ‘data dictionary’ to
understand all the meanings of the columns (e.g. what is “plot type” in
ratdat?) We don’t our helper to waste valuable time to figure out what everything means. - highly derived/modified from the original file. You may have already done a bunch of preliminary data wrangling you don’t want to include when you send the example, so you would need to provide the intermediate dataset directly to your helper.
While Mickey does not have to create a brand new example dataset, they should at least work to make their original data minimal and reproducible (see the reflection exercise above).
While it may sometimes feel like unnecessary effort, the process of creating a minimal dataset will not only help others help you, but also allow you to better understand your data and often discover the source of the problem without the need for external help. By removing extraneous information and only keeping what is required to replicate the issue, we can ensure both we and our helper are be able to easily see how the data is structured and where the problem arises. Furthermore, we have already seen how sometimes the source of the problem isn’t actually the code, but rather the data! Providing an appropriate example, or mock dataset allows a helper to better investigate and manipulate that data to fix the problem (as if they were working directly on your computer).
See what the mock dataset of a reprex can look like in the callout below.
While formatted as a callout, the pro-tip content is critical for students to develop the right mental model of what an example dataset looks like.
Pro-tip: documentation examples are a reprex
An example of what minimal reproducible examples look like can be
found in the ?help section, in R Studio. When looking at
the documentation for any function, scroll all the way down to where
they list examples. These will usually be minimal and reproducible,
since they are intended to be directly copy-pasted and run by
anyone.
For example, let’s look at the function mean:
R
?mean
When we scroll all the way down, we see examples that can be run directly on the console, with no additional code. Try to copy-paste the following in your script. Does it run?
R
xm <- mean(x)
ERROR
Error: object 'x' not foundR
c(xm, mean(x, trim = 0.10))
ERROR
Error: object 'xm' not foundIt is missing the first line which actually creates the dataset
x. Try again with the new line.
R
x <- c(0:10, 50)
xm <- mean(x)
c(xm, mean(x, trim = 0.10))
OUTPUT
[1] 8.75 5.50Now it should run as intended!
In this example, x is the mock dataset consisting of just 1 variable. Notice how it was created as part of the example. This will be your goal with your reprex.
When your data is uploaded from a separate file, that file becomes a dependency in your code–it cannot run without it. Since a reprex needs to be reproducible by anyone, we need to think about our data in a different way–as a mock object created in the script itself.
In summary, just like with the code, a minimal reproducible dataset must be:
- minimal: it only contains information required to run your minimal code and replicate the problem. You can also think of this as being relevant to the problem.
- reproducible: it must be accessible to someone without your computer, and it must consistently replicate your problem. This means it also needs to be complete, meaning there are no dependencies that have been omitted (e.g., packages).
This exercise is intended to address the LO “Describe a minimal reproducible dataset” although it better targets the learner’s ability to identify an appropriate reprex dataset.
Ecercise 3: Test your knowledge!
Let’s say we want to know the average weight of all the species in
our rodents dataset from episode 2 (also below if you no
longer have it). We try to use the following code…
R
# The previously created "rodents" dataset
rodents <- surveys %>% filter(taxa == "Rodent")
# Mean rodent weight
mean(rodents$weight)
OUTPUT
[1] NA…but it returns NA! We don’t know why that is happening and we want to ask for help.
Which of the following represents a minimal reproducible dataset for
the minimal code mean(rodents$weight)? Can you describe why
the other ones are not?
sample_data <- data.frame(month = rep(7:9, each = 2), hindfoot_length = c(10, 25, 14, 26, 30, 17))sample_data <- data.frame(weight = rnorm(10))sample_data <- data.frame(weight = c(100, NA, 30, 60, 40, NA))sample_data <- rodents_modified[1:20,]
Hint: the data needs to (1) run within your code,
and (2) replicate the problem. To test whether each new
sample_data works, try inputting it back into the function
that is giving you problems. E.g., use
mean(sample_data$weight)
The correct answer is C!
- does not include the variable of interest (weight).
- does not replicate the problem (NA result with a warning message)–the code runs just fine.
- minimal and reproducible.
- is not reproducible. Sample randomly samples 10 items; sometimes it may include NAs, sometime it may not (not guaranteed to replicate the error). It can be used if a seed is set (see next section for more info).
- uses a dataset that isn’t accessible without previous data wrangling code–the object rodents_modified doesn’t exist.
- Inaccessible data. It provides the path to a file on someone else’s computer.
Don’t use screenshots
Screenshots are another example of an inappropriate and unhelpful way to share your data with a helper. While they can be used as a quick snapshot of what the data looks like, the data within a screenshot cannot be manipulated and a helper would have to create their own mock dataset to run the code.
4.2 Three different approaches
In general, there are 3 common ways to provide minimal reproducible data for a reprex.
Add a few lines of code to create a mock dataset with the same key characteristics as the original data (like in the example sections of documentations).
Subset the original data to be minimal and reproducible, then use function like
dput().Subset a dataset that is already provided by R (e.g.,
cars,npk,penguins, etc.). For a complete list, uselibrary(help = "datasets").
It would be nice if we could toggle the callout below or at least the table so that it doesn’t take so much room since it is not an essential part of the lesson.
Pros and Cons
The developers of this lesson believe everyone is entitled to use any option they prefer, therefore the rest of this episode will expand on each of the 3 approaches listed above. However, within the data science community, opinions differ on which method is best recommended. Below is a summary table of advantages and disadvantages of each approach based on many conversations with several data science groups.
| Advantages | Disadvantages | |
| Data from Scratch |
|
|
| R-built Data |
|
|
| Your Data |
|
|
4.3 Creating a mock dataset from scratch
While starting from scratch can be daunting at first, it becomes easier and faster with practice. Once you are familiar with the basic building blocks, it is a straight-forward method of creating a minimal reproducible dataset. This is also the preferred method for other activities that require a reprex (e.g., teaching, collaborating, developing, etc.), and it often provides valuable problem-solving insights. So let’s breakdown this process to be more digestible!
Mickey is still new at this and has 2 pressing questions:
- How do I create a dataset from scratch?
- How do I know which key aspects of my data to recreate?
Let’s start with the first.
There are many ways one can create a dataset in R (these should be familiar if you took the Carpentries lesson Data Analysis and Visualization in R for Ecologists).
You can start by creating vectors using c()
R
vector <- c(1,2,3,4)
vector
OUTPUT
[1] 1 2 3 4You can also add some randomness by sampling from a vector using
sample().
For example you can sample numbers 1 through 10 in a random order
R
x <- sample(1:10, 5)
x
OUTPUT
[1] 9 1 2 6 7Or you can randomly sample from a normal distribution
R
x <- rnorm(10)
x
OUTPUT
[1] -1.09661389 -0.38641709 -0.02238582 0.11590507 -0.49023878 -1.33091769
[7] -0.03217919 -0.30498972 0.53778249 -0.78195417You can also use built-in vectors like letters to create
factors.
R
x <- sample(letters[1:4], 20, replace=T)
x
OUTPUT
[1] "c" "b" "c" "a" "a" "c" "d" "b" "d" "b" "b" "c" "c" "b" "c" "c" "a" "b" "c"
[20] "d"Remember that a data frame is just a collection of
vectors. You can create a data frame using
data.frame (or tibble in the
dplyr package), and then define a vector for each
variable.
R
data <- data.frame (x = sample(letters[1:3], 20, replace=T),
y = rnorm(1:20))
head(data)
OUTPUT
x y
1 b 1.7434602
2 c -1.0455049
3 c 0.1794809
4 c -1.4669104
5 b -0.3858596
6 c 0.2545273However, when sampling at random you must remember
to set.seed() before sending it to someone to make sure you
both get the same numbers!
For more handy functions for creating data frames and variables, see
the [cheatsheet]. Some questions may require specific
formats. For these, you can use any of the provided
as.someType functions: as.factor,
as.integer, as.numeric,
as.character, as.Date,
as.xts.
You can skip the challenge below if short on time. The exercise assesses the LO: “Create a dataset from scratch.” There will not be another exercise to practice this, but you will walk through creating the main reprex dataset together, so that may be enough. Do not spend too much time on this. Students can work in pairs, or you can work all together. Don’t wait for everyone to be done before going through solutions, maybe have them alert you when they finish each one, they can then keep going while they wait. You can skip giving the solution for C, expecting quicker learners to have tried it for practice while slower learners may not have gotten to it, that’s ok!
Exercise 4: You try!
Create a data frame with:
A. One categorical variable with 2 levels and one continuous variable.
B. One continuous variable that is normally distributed.
C. Name, sex, age, and treatment type.
4.5 Identifying the relevant aspects of your data
No matter which approach you choose to take for providing a dataset, they key is always to identify which elements of the original data are necessary, or relevant, to replicate the problem. To do so, here are a few guiding questions:
- Which variables are necessary to replicate the problem?
- What data type (discrete or continuous) is each variable?
- How many levels and/or observations are necessary?
- Do the values need to be distributed in a specific way?
- Are there any NAs that could be relevant?
Let’s check back with Mickey and the minimal code they settled on:
R
# Mickey's current minimal code
library(readr)
library(dplyr)
surveys <- read_csv("data/surveys_complete_77_89.csv")
OUTPUT
Rows: 16878 Columns: 13
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (6): species_id, sex, genus, species, taxa, plot_type
dbl (7): record_id, month, day, year, plot_id, hindfoot_length, weight
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.R
# Filter to known sex
rodents_subset <- surveys %>%
filter(sex == c("F", "M"))
# Subsetted dataset
table(rodents_subset$sex)
OUTPUT
F M
3656 4145 R
# Original dataset
table(surveys$sex) # the numbers don't match!
OUTPUT
F M
7318 8260 Let’s take a closer look at the dataset we need to substitute and then answer the questions outlined earlier.
R
head(surveys)
OUTPUT
# A tibble: 6 × 13
record_id month day year plot_id species_id sex hindfoot_length weight
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl>
1 1 7 16 1977 2 NL M 32 NA
2 2 7 16 1977 3 NL M 33 NA
3 3 7 16 1977 2 DM F 37 NA
4 4 7 16 1977 7 DM M 36 NA
5 5 7 16 1977 3 DM M 35 NA
6 6 7 16 1977 1 PF M 14 NA
# ℹ 4 more variables: genus <chr>, species <chr>, taxa <chr>, plot_type <chr>Exercise 5: Your turn!
Try to answer the following questions on your own to determine what we need to include in our minimal reproducible dataset:
- Which variables does Mickey need to replicate their problem?
- What data type (discrete or continuous) is each variable?
- How many levels and/or observations are necessary?
- Do the values need to be distributed in a specific way?
- Are there any NAs that could be relevant?
Give them some time to think it through, maybe in pairs or small groups, but focus on working through the answers together, even if not everyone has finished. Go one question at a time (time to answer the first -> walk through solution -> time to answer the second -> etc.).
Let’s go over the answers together and help Mickey build a dataset as we go along!
Begin creating the dataset with the information gained from each question, then you can put it all together.
- How many variables does Mickey need to reproduce their problem?
They really just sex, and maybe an identifier like record_id. This means they potentially only need 1 vector, maybe 2 (remember, each column in a dataframe is essentially a vector, and in “tidy data” should correspond with a variable; each row is then an observation).
R
# create 2 variables: sex, and maybe record_id
# a vector for sex:
# a vector for record_id:
- What data type (discrete or continuous) is each variable?
Sex is a discrete (categorical) variable, while record ID would be continuous.
R
# create 2 variables: sex, and maybe record_id
# a vector for sex: categorical
# a vector for record_id: continuous
- How many levels and/or observations are necessary?
Since Mickey is filtering their dataset down to 2 categories for sex, that means they need at least 3 levels to start with. In terms of number of observations there don’t seem to be specific restrictions so they can just pick a generally nice number like 10. This is where creating a reprex dataset becomes a bit more of an art than a science; it is common to use trial and error until the problem is replicated accurately.
R
# create 2 variables: sex, and maybe record_id
# a vector for sex: categorical with 3 levels
# a vector for record_id: continuous, ~10
- Do the values need to be distributed in a specific way?
This question probably isn’t going to be relevant most of the time, but certainly worth considering. If Mickey needed a longer dataset of measurements they may have wanted to make sure it was normally distributed. If they needed a longer dataset of counts they may have wanted to make sure it was Poisson distributed. Or maybe they had binary data. But in this case, Mickey has a fairly short dataset and the code doesn’t include anything that should vary depending on the distribution, so it probably doesn’t matter. Again, this process can be one of trial and error. They can always come back to this question if they are unable to replicate their problem (hint: in which case the distribution may be related to the problem they are having!).
- Are there any NAs that could be relevant?
Mickey’s data does have NAs for the sex variable. It might not matter or it could be important, so let’s have them put in NAs in the mock dataset just in case.
R
# create 2 variables: sex, and maybe record_id
# a vector for sex: categorical with 3 levels, one of which is NA
sex <- c('M','F',NA)
sex
OUTPUT
[1] "M" "F" NA R
# a vector for record_id: continuous, ~10
record_id <- 1:10
record_id
OUTPUT
[1] 1 2 3 4 5 6 7 8 9 10R
# Now let's go "sampling" and put our "obervations" in a dataframe
sample_data <- data.frame(
# record_id stays the same, since these are our 10 "observations"
record_id = record_id,
# randomly select 10 observations from our list of sexes
sex = sample(sex, 10, replace=T)
)
# Look at our new dataset
sample_data
OUTPUT
record_id sex
1 1 <NA>
2 2 <NA>
3 3 <NA>
4 4 F
5 5 M
6 6 M
7 7 M
8 8 F
9 9 <NA>
10 10 FAnd just like that we helped Mickey create a mock dataset from
scratch! Notice that they could also have compiled the same type of
dataset in a single line by creating each vector within
data.frame()
R
sample2_data <- data.frame(
record_id = 1:10,
sex = sample(c('M','F', NA), 10, replace=T)
)
sample2_data
OUTPUT
record_id sex
1 1 F
2 2 <NA>
3 3 F
4 4 F
5 5 F
6 6 M
7 7 M
8 8 M
9 9 M
10 10 <NA>Important: Notice that the outputs of the two
datasets are not the same. If you want the outputs to be EXACTLY the
same each time, but you are using sample() which is an
inherently random process, you must first use set.seed()
and share that with your helper too.
R
set.seed(1) # set seed before recreating the sample
sample_data <- data.frame(
record_id = 1:10,
sex = sample(c('M','F', NA), 10, replace=T)
)
sample_data
OUTPUT
record_id sex
1 1 M
2 2 <NA>
3 3 M
4 4 F
5 5 M
6 6 <NA>
7 7 <NA>
8 8 F
9 9 F
10 10 <NA>Adding a set.seed() at the start of your reprex will
ensure anyone else who runs the same code in the same
order will always get the same results. However, if using it
more generally, you may want to read more about it. For example, in the
example below we set a seed of 2 and then run sample(10)
twice. You will notice that the output of each sample run is not the
same. However, if you run the whole code again, you will see that each
of the outputs actually do stay the same.
R
set.seed(2)
sample(10)
OUTPUT
[1] 5 6 9 1 10 7 4 8 3 2R
sample(10)
OUTPUT
[1] 1 3 6 2 9 10 7 5 4 8Great! Now we need to check whether the mock dataset works with the minimal code Mickey created earlier. Does it run? Does it replicate the problem they were having?
R
# Mickey's reprex (1 approach)
# Required packages to run the code
library(readr)
library(dplyr)
set.seed(1) # ensures accurate data replication
# Create a mock dataset
sample_data <- data.frame(
record_id = 1:10,
sex = sample(c('M','F', NA), 10, replace=T)
)
# The problematic code snippet
sample_subset <- sample_data %>% # replace rodents with our sample dataset
filter(sex == c("F", "M")) # this can stay the same because we recreated it the same
# Subsetted sample dataset - how many individuals for each sex?
table(sample_subset$sex)
OUTPUT
F
1 R
# Original sample dataset - how many individuals for each sex?
table(sample_data$sex) # the numbers don't match!
OUTPUT
F M
3 3 It works! The sample size has unexpectedly been reduced to just 2 observations, when we would have expected a sample of 8, based on the sample_data output above. Wherever the issue may lie, we were able to successfully replicate it in this minimal reproducible example.
4.6 Using the original data set
Even if you master the art of creating mock datasets, there may be occasions in which your data or problem is too complex and you can’t seem to replicate the issue. Or maybe you still think using your original data would just be easier.
In cases when you want to make your own data minimal and reproducible, you will want to take a similar approach to what we did in Episode 3 when making the code minimal. Keep what is essential, get rid of the rest. In other words, we will want to subset our data into a smaller, more digestible chunk.
The question still arises: how do I know what is essential?
Use the same guiding questions that we used earlier!
- Which variables are necessary to replicate the problem?
- What data type (discrete or continuous) is each variable? (perhaps less necessary, since you are keeping the original variables)
- How many levels and/or observations are necessary? (we don’t want to get rid of more than we need)
- Do the values need to be distributed in a specific way? (worth keeping in mind in terms of how we are removing observations)
- Are there any NAs that could be relevant?
You can give them time to answer those questions on their own first, like in Exercise 5, or just go through them together.
Based on our previous answers we end up with:
- We need species, sex, and maybe record_id
- Species and sex are categorical, record_id is a continuous count of our observations.
- As we said earlier, we want 3 each for species and sex, which happens to already be the case. And we could reduce our record_id size to ~10.
- Not really, but we want to make sure that when we reduce the number of observations we still have observations in each of the 3 levels in species and sex.
- NA’s are present in the sex variable, so let’s make sure we keep at least one.
Now that we have a clearer goal, let’s subset the data.
Useful functions for subsetting a dataset include
subset(), head(), tail(), and
indexing with [] (e.g., iris[1:4,]). Alternatively, you can use
tidyverse functions like select(), and
filter() from the tidyverse. You can also use the same
sample() functions we covered earlier.
Note: you should already have an understanding of how to subset or wrangle data using the tidyverse from the Data Analysis and Visualization in R for Ecologists. If not, go check it out!
R
# Mickey's current script
library(readr)
library(dplyr)
surveys <- read_csv("data/surveys_complete_77_89.csv")
OUTPUT
Rows: 16878 Columns: 13
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (6): species_id, sex, genus, species, taxa, plot_type
dbl (7): record_id, month, day, year, plot_id, hindfoot_length, weight
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.R
# Filter to known sex
rodents_subset <- surveys %>%
filter(sex == c("F", "M"))
# Subsetted dataset
table(rodents_subset$sex)
OUTPUT
F M
3656 4145 R
# Original dataset
table(surveys$sex) # the numbers don't match!
OUTPUT
F M
7318 8260 Given that the code that is going wrong is that which creates rodents_subset, we need to create a minimal reproducible version of surveys! We can then insert our new_surveys dataset in place of the original rodents one.
Step 1: select the variables of interest
R
# subset rodent into new_rodent to make it minimal
# Note: there are many ways you could do this!
new_surveys <- surveys %>%
# 1. select the variables of interest
select(record_id, sex)
# PAUSE. Does this work so far?
new_surveys
OUTPUT
# A tibble: 16,878 × 2
record_id sex
<dbl> <chr>
1 1 M
2 2 M
3 3 F
4 4 M
5 5 M
6 6 M
7 7 F
8 8 M
9 9 F
10 10 F
# ℹ 16,868 more rowsStep 2-5: reduce the number of observations to ~10 while making sure the dataset still contains at least 3 species and at least 3 sexes
While the rest is just one step, it is the trickiest, because this is where we want to ensure the key elements of our original dataset, as defined earlier, are preserved.
Exercise 6: Your Turn! (5 mins)
How would you continue the subsetting pipeline? How could you reduce the number of observations while making sure you still have at least 3 sexes left? Hint: there is no single right answer! Trial and error works wonders.
R
set.seed(1)
new_surveys <- surveys %>%
# select the variables of interest
select(record_id, sex) %>%
# randomly select 4 rows from each sex category
slice_sample(n=4, replace = F, by='sex') # use ?slice_sample to check the documentation
new_surveys
OUTPUT
# A tibble: 12 × 2
record_id sex
<dbl> <chr>
1 2359 M
2 16335 M
3 9910 M
4 8278 M
5 12038 F
6 7862 F
7 9221 F
8 1335 F
9 3320 <NA>
10 343 <NA>
11 14482 <NA>
12 9376 <NA> The code ran without issues, hooray! But do we end up with what we were looking for?
- Do we have ~10 observations? Yes! 12 sounds good
- Do we have at least 3 sexes? Yes! M, F, and NA
Great! All of our requirements are fulfilled. Now let’s see if it replicates Mickey’s problem when we add it to their minimal code.
Note: slice_sample() and
similar functions allow you to specify and customize how exactly you
want that sample to be taken (check the documentation!). For example,
you can specify a proportion of rows to select, specify how to order
variables, whether ties [may require more explanation]
should be kept together, or even whether to weigh certain variables. All
of this allows you to keep aspects of your dataset that may be relevant
and hard to replicate otherwise.
Remember the problematic code snippet:
R
# Filter to known sex
rodents_subset <- surveys %>%
filter(sex == c("F", "M"))
We now want replace surveys with
new_surveys, and let’s change the name of the
rodents_subset too so we can compare them. Does that
problematic code snippet work or do we need to change anything else?
R
# Filter to known sex
new_subset <- new_surveys %>%
filter(sex == c("F", "M"))
The code ran without any errors! But does it replicate Mickey’s problem?
Note: if the new dataset had an odd number of rows it would spit out a warning giving you a hint as to what the source of the problem may be.
Take a step back to remind yourself of what you are looking for. What was the problem we had identified?
- The number of rows that remain after the filter is lower than expected.
So what would we expect to see with this new dataset? Since it is nice and short, this makes it a lot easier to predict the outcome.
- We should find that both tables list 4 females and 4 males.
R
# Before the filter
table(new_surveys$sex) # as expected!
OUTPUT
F M
4 4 R
# After the filter
table(new_subset$sex) # oh no! The numbers listed are not what we expect...
OUTPUT
F M
2 2 R
# ...which is exactly the problem we were trying to replicate!
Why aren’t we getting the rows we are asking for?
Maybe our table is just wrong, let’s look at the actual dataset we end up with
R
new_subset
OUTPUT
# A tibble: 4 × 2
record_id sex
<dbl> <chr>
1 16335 M
2 8278 M
3 12038 F
4 9221 F Still wrong! What is going on?? We don’t have an answer, but we certainly replicated a problem that occurs when we filter the data. Time to ask for help!
I love that the filter {dplyr} documentation is
thoroughly unhelpful on this
But wait, Mickey’s dataset is now minimal and relevant, but is it
reproducible (accessible outside their device)? Not yet. We created a
subset of their original dataset rodents but this came from
a file on their computer. They could share the csv file and add a line
of code that uploads it… but we already said this is not good practice
for a reprex, and it makes it hard to ask for help on a community
website. Remember, the more steps required, the less likely someone will
want to help.
Thankfully, there is a nifty function dput() that can
help us out. Let’s try it and see what happens.
R
dput(new_surveys)
OUTPUT
structure(list(record_id = c(2359, 16335, 9910, 8278, 12038,
7862, 9221, 1335, 3320, 343, 14482, 9376), sex = c("M", "M",
"M", "M", "F", "F", "F", "F", NA, NA, NA, NA)), row.names = c(NA,
-12L), class = c("tbl_df", "tbl", "data.frame"))It spit out a hard-to-read but not excessively long chunk of code.
This code, when run, will recreate the new_rodents dataset!
We can also break it down and label it further to help the reader.
R
reprex_data <- structure(list(
# a unique identifier (not actually necessary)
record_id = c(2359, 16335, 9910, 8278, 12038,
7862, 9221, 1335, 3320, 343, 14482, 9376),
# A list of sexes. Note: this includes NAs!
sex = c("M", "M",
"M", "M", "F", "F", "F", "F", NA, NA, NA, NA)),
# no row names
row.names = c(NA, -12L),
# type of data
class = c("tbl_df", "tbl", "data.frame"))
# see the dataset
print(reprex_data)
OUTPUT
# A tibble: 12 × 2
record_id sex
<dbl> <chr>
1 2359 M
2 16335 M
3 9910 M
4 8278 M
5 12038 F
6 7862 F
7 9221 F
8 1335 F
9 3320 <NA>
10 343 <NA>
11 14482 <NA>
12 9376 <NA> Ta-da! Now anyone can easily recreate Mickey’s minimal dataset and use it to run the minimal code. Now, was that really easier than creating a dataset from scratch?
Some of you may still be thinking that you could just use
dput() on the original dataset. And it would work. But that
wouldn’t be very considerate to those who are trying to help.
Exercise 7: Try it! (1 min)
Try running dput(surveys) in your script.
It becomes a huge chunk of code! When clearly we don’t need all of that.
Remember, we want to keep everything minimal for many reasons:
- to make it easy for our helpers to understand our data and code
- to allow helpers to quickly focus their efforts on the right factors
- to make the problem-solving process as easy and painless as possible
- bonus: to help us better understand and zero-in on the source of our problem, often stumbling upon a solution along the way
Nevertheless, it remains an option for when your data appears too complex or you are not quite sure where your problem lies and therefore are not sure what minimal components are needed to reproduce the example. In other words, when you don’t have a good mental model of what the problem is even after going through the initial steps we outlined earlier in the lesson.
4.7 Using an R-built dataset
The last approach we mentioned is to build a minimal reproducible dataset based on the datasets that already exist within R (and therefore everyone would have access to).
A list of readily available datasets can be found using
library(help="datasets"). You can then use ?
in front of the dataset name to get more information about the contents
of the dataset.
For a more detailed discussion of the benefits of using this approach see the Pros and Cons callout in section 4.3.
This approach essentially blends the skills you already learned in
the first two. You need to identify a dataset with appropriate variables
that match the “key elements” of the original dataset. You then need to
further reduce that dataset to a minimal, relevant, number or rows. Once
again, you can use the previously learned functions such as
select(), filter(), or
sample().
Since you already learned everything you need, why not try it yourself?
Exercise 8: Extra Challenge (10 mins)
Using the “HairEyeColor” dataset, create a minimal reproducible dataset for the same issue and minimal code we have been exploring.
Start by using
?HairEyeColorto read a description of the dataset andView(HairEyeColor)to see the actual dataset.Which variables would be a good match for our situation? What are our requirements?
How can we subset this dataset to make it minimal and still replicate our issue?
Remember, there are many possible solutions! The most important feature is that the example dataset can replicate the issue when used within our minimal code.
The following is 1 possible solution:
We selected Hair and Eye as replacements for species and sex because
they are both categorical and have at least 3 levels. We don’t strictly
need anything else. We will call our new survyes
replacement hyc. We set a seed because we want a random
sample.
R
set.seed(1)
# the dummy dataset
hyc <- as.data.frame(HairEyeColor) %>% # oh no! Needs to be converted to df -- might need to change example or have them figure that one out... or we can give them this first line.
select(Hair, Eye) %>%
slice_sample(n=10)
print(hyc)
OUTPUT
Hair Eye
1 Black Hazel
2 Blond Brown
3 Red Blue
4 Black Brown
5 Brown Brown
6 Red Blue
7 Red Hazel
8 Brown Green
9 Brown Brown
10 Red BrownR
# the minimal code
hyc_subset <- hyc %>%
filter(Hair == c('Red','Blonde'),
Eye == c('Blue', 'Brown'))
# illustrating the issue
table(hyc_subset$Hair, hyc_subset$Eye)
OUTPUT
Brown Blue Hazel Green
Black 0 0 0 0
Brown 0 0 0 0
Red 0 1 0 0
Blond 0 0 0 0R
# the whole subset
print(hyc_subset)
OUTPUT
Hair Eye
1 Red BlueR
# But we know there are more!
table(hyc$Hair, hyc$Eye) # Reds have 2 Blue and 1 Brown, and Blonds have 1 Brown!
OUTPUT
Brown Blue Hazel Green
Black 1 0 1 0
Brown 2 0 0 1
Red 1 2 1 0
Blond 1 0 0 0What about NAs?
If your data has NAs and they may be causing the problem, it is
important to include them in your example dataset. You can find where
there are NAs in your dataset by using is.na, for example:
is.na(krats$weight). This will return a logical vector or
TRUE if the cell contains an NA and FALSE if not. The simplest way to
include NAs in your dummy dataset is to directly include it in vectors:
x <- c(1,2,3,NA). You can also subset a dataset that
already contains NAs, or change some of the values to NAs using
mutate(ifelse()) or substitute all the values in a column
by sampling from within a vector that contains NAs.
One important thing to note when subsetting a dataset with NAs is that subsetting methods that use a condition to match rows won’t necessarily match NA values in the way you expect. For example
R
test <- data.frame(x = c(NA, NA, 3, 1), y = rnorm(4))
test %>% filter(x != 3)
OUTPUT
x y
1 1 -0.3053884R
# you might expect that the NA values would be included, since “NA is not equal to 3”. But actually, the expression NA != 3 evaluates to NA, not TRUE. So the NA rows will be dropped!
# Instead you should use is.na() to match NAs
test %>% filter(x != 3 | is.na(x))
OUTPUT
x y
1 NA 0.4874291
2 NA 0.7383247
3 1 -0.3053884Great work! You created a minimal reproducible example. In the next episode, you will learn about reprex, a package that can help you double-check that your example is properly reproducible by running it in a clean environment. (As an added bonus, reprex will format your example nicely so it’s easy to post to places like Slack, GitHub, and StackOverflow.)
- A minimal reproducible dataset (a) contains the minimum number of lines, variables, and categories, in the correct format, to replicate your problem; and (b) must be fully reproducible, meaning that someone else can run the same code from anywhere without additional steps.
- To make it accessible, you can create a dataset from scratch using
as.data.frame, you can use an R-built dataset likecars, or you can use a subset of your own dataset and then usedput()to generate reproducible code.
Bonus: Additional Practice
Here are some more practice exercises if you wish to test your knowledge
Extra Practice? Would need to change from mpg, since that’s from ggplot
For each of the following, identify which data are necessary to
create a minimal reproducible dataset using mpg.
- We want to know how the highway mpg has changed over the years
- We need a list of all “types” of cars and their fuel type for each
manufacturer
- We want to compare the average city mpg for a compact car from each
manufacturer
(I copied these from excercise 6 in the google doc… but I’m not sure that they are getting at the point of the lesson…)
Another Excercise
Each of the following examples needs your help to create a dataset that will correctly reproduce the given result and/or warning message when the code is run. Fix the dataset shown or fill in the blanks so it reproduces the problem.
-
set.seed(1)sample_data <- data.frame(fruit = rep(c(“apple”, “banana”), 6), weight = rnorm(12))ggplot(sample_data, aes(x = fruit, y = weight)) + geom_boxplot()HELP: how do I insert an image from clipboard?? Is it even possible? - bodyweight <- c(12, 13, 14, , ) max(bodyweight) [1] NA
- sample_data <- data.frame(x = 1:3, y = 4:6) mean(sample_data\(x) [1] NA Warning message: In mean.default(sample_data\)x): argument is not numeric or logical: returning NA
- sample_data <- ____ dim(sample_data) NULL
- “fruit” needs to be a factor and the order of the levels must be
specified:
sample_data <- data.frame(fruit = factor(rep(c("apple", "banana"), 6), levels = c("banana", "apple")), weight = rnorm(12)) - one of the blanks must be an NA
- ?? + what’s really the point of this one?
sample_data <- data.frame(x = factor(1:3), y = 4:6)
At the end of the episode, here is our final reprex. In the next episode, we will learn how to test whether this reprex is fully reproducible, and also how to share it with others.
R
# Mickey's reprex (1 approach)
# Required packages to run the code
library(readr)
library(dplyr)
set.seed(1) # ensures accurate data replication
# Create a mock dataset
sample_data <- data.frame(
record_id = 1:10,
sex = sample(c('M','F', NA), 10, replace=T)
)
# The problematic code snippet
sample_subset <- sample_data %>% # replace rodents with our sample dataset
filter(sex == c("F", "M")) # this can stay the same because we recreated it the same
# Subsetted sample dataset - how many individuals for each sex?
table(sample_subset$sex)
OUTPUT
F
1 Content from Asking your question
Last updated on 2026-02-24 | Edit this page
Estimated time: 12 minutes
Overview
Questions
- How can I verify that my example is reproducible?
- How can I easily share a reproducible example with a mentor or helper, or online?
- What information should I include when asking a question?
Objectives
- Use the {reprex} package to test whether an example is reproducible.
- Use the {reprex} package to format reprexes for posting online.
- Understand the benefits and drawbacks of different help forums.
- Have a road map to follow when posting a question to make sure it’s a good question.
- Understand what the {reprex} package does and doesn’t do.
In the previous episodes, we have worked on creating a minimal reproducible example, also known as a “reprex”, to help our researcher friend Mickey debug a problem with their code.
We learned how to 1) identify the problem and try some preliminary fixes, 2) minimize the code, and 3) include a minimal dataset.
In this episode, we will introduce a tool, the reprex package, that we can use to verify that we’ve successfully created a minimal reproducible example. The {reprex} package will also help format the code nicely for posting online or sending to a colleague.
When preparing to share a reprex with other people, it is important to remember three principles:
- Reproducibility
- Formatting
- Context
1. Reproducibility
Haven’t we already talked a lot about reproducibility?
Yes! We have discussed variables and packages, minimal datasets, and making sure that the problem is meaningfully replicated by the data that you choose. But there are some reasons that a code snippet that appears reproducible in your own R session might not actually be runnable by someone else.
You forgot to account for the origins of some functions and/or variables. We went through our code methodically, but what if we missed something? It would be nice to confirm that the code is as self-contained as we thought it was.
Your code depends on some particular characteristic of your R or RStudio environment that is not the same as your helper’s environment. For example, you might be using an older version of a package, and the problem could be fixed by reinstalling a newer version. Another package you have loaded could have a function with a conflicting name. Maybe something is up with your operating system, time zone, R version, or RStudio preferences that a helper can’t necessarily replicate.
Checking all of these possible causes of errors feels overwhelming, and it may not be obvious how to do it. Even experienced R coders do not always feel comfortable changing their RStudio settings, managing package versions, or accessing hidden configuration files.
Especially for complex problems, it is useful to have a way to double check that our painstakingly-prepared reprexes are actually minimal and reproducible, without manually testing out every contingency in R!
Luckily, the {reprex} package will help you test your reprexes in a clean, isolated environment to make sure they’re actually reproducible.
The most important function in the reprex package is
called reprex(), and using it looks a little different from
functions in many other packages. reprex() automatically
recognizes code that you have copied to your computer’s
clipboard and uses it to create a nicely formatted
reprex, which it puts onto the clipboard for you to paste elsewhere.
Because this happens invisibly, it can be a little hard to keep track
of. Here’s what’s happening behind the scenes.
[DIAGRAM OF THE CLIPBOARD HERE]
Your computer uses the clipboard every time you copy and paste
something! What makes reprex() different is that it
automatically reads and writes from the keyboard; you don’t have to
manually paste the code into the reprex() function.
Let’s try it out. We’ll first create a very minimal reproducible example, such as this one from the reprex package documentation.
R
y <- 1:4
mean(y)
OUTPUT
[1] 2.5To turn this code into a reprex, we can highlight the code and copy
it to your clipboard. (e.g. Cmd + C on Mac, or
Ctrl + C on Windows).
Finally, in your console, type reprex(). Leave the
parentheses empty. You don’t have to paste anything. Hit
Enter/Return.
# (with the target code snippet copied to your clipboard already...)
# In the console:
reprex()reprex will grab the code that you copied to your
clipboard and run that code in an isolated environment. It will
return a nicely formatted reproducible example that includes your code
and and any results, plots, warnings, or errors generated.
Callout: The reprex package
workflow
The reprex package workflow takes some getting used to.
Instead of copying your code into the function, you simply copy
it to the clipboard (a mysterious, invisible place to most of us) and
then let the blank, empty reprex() function go over to the
clipboard by itself and find it.
And then the completed, rendered reprex replaces the original code on the clipboard and all you need to do is paste, not copy and paste.
Let’s practice this one more time. Here’s some code that makes a boxplot:
R
library(ggplot2)
library(dplyr)
mpg %>%
ggplot(aes(x = factor(cyl), y = displ))+
geom_boxplot()
Once again, we can highlight the code snippet, copy it to the
clipboard, and then run reprex() in the console.
# In the console:
reprex()The result, which was automatically placed onto my clipboard and which I pasted here, looks like this:
R
library(ggplot2)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
mpg %>%
ggplot(aes(x = factor(cyl), y = displ))+
geom_boxplot()

Created on 2024-12-29 with reprex v2.1.1
Nice and neat! It even includes the plot produced, so I don’t have to take screenshots and figure out how to attach them to an email.
{reprex} as a debugging collaborator
The formatting is great, but {reprex} really shines when you treat it as a helpful collaborator in your process of building a reproducible example.
Exercise 1: Using {reprex} to find problems with a minimal reproducible example
If you run the code from the previous example without running the
library(ggplot2)line, does it still work? Why?Make a prediction: What will happen if you forget to include
library(ggplot2)in your minimal reproducible example? Try it with the {reprex} package and see. Why did this happen? ::: solution
R
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
mpg %>%
ggplot(aes(x = factor(cyl), y = displ))+
geom_boxplot()
#> Error in ggplot(., aes(x = factor(cyl), y = displ)): could not find function "ggplot"
Created on 2024-12-29 with reprex v2.1.1
:::
We get an error message indicating that R cannot find the function
ggplot! That’s because we forgot to load the
ggplot2 package in the reprex.
This happened even though we had ggplot2 already loaded
in our own current RStudio session. reprex deliberately
ignores any packages already loaded, running the code in a clean,
isolated R session that’s different from the R session we’ve been
working in. This simulates the experience of someone else trying to
run your reprex on their own computer.
Testing it out
Now that we’ve met our new reprex-making collaborator, let’s use it to test out the reproducible example we created in the previous episode.
Here’s the code we wrote:
R
# Mickey's reprex (1 approach)
# Required packages to run the code
library(readr)
library(dplyr)
set.seed(1) # ensures accurate data replication
# Create a mock dataset
sample_data <- data.frame(
record_id = 1:10,
sex = sample(c('M','F', NA), 10, replace=T)
)
# The problematic code snippet
sample_subset <- sample_data %>%
filter(sex == c("F", "M"))
# Subsetted sample dataset - how many individuals for each sex?
table(sample_subset$sex)
It’s time to verify if our example is actually reproducible! Let’s
copy it to the clipboard and run reprex().
# In the console:
reprex()It worked!
R
# Mickey's reprex (1 approach)
# Required packages to run the code
library(readr)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
set.seed(1) # ensures accurate data replication
# Create a mock dataset
sample_data <- data.frame(
record_id = 1:10,
sex = sample(c('M','F', NA), 10, replace=T)
)
# The problematic code snippet
sample_subset <- sample_data %>%
filter(sex == c("F", "M"))
# Subsetted sample dataset - how many individuals for each sex?
table(sample_subset$sex)
#>
#> F
#> 1
Created on 2025-09-11 with reprex v2.1.1
Now we have a beautifully-formatted reprex that includes runnable code and all the context needed to reproduce the problem.
This also would have been a good way for Mickey to discover that they needed to create minimal data earlier! Look what would have happened if we had put the example from the end of episode 3 into {reprex}:
R
library(readr)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
surveys <- read_csv("data/surveys_complete_77_89.csv")
#> Error: 'data/surveys_complete_77_89.csv' does not exist in current working directory ('/private/var/folders/xy/lqbp515s27q8v1drkll9gynw0000gr/T/RtmpJX7W20/reprex-76216582aaa-irate-booby').
# Filter to known sex
rodents_subset <- surveys %>%
filter(sex == c("F", "M"))
#> Error: object 'surveys' not found
# Subsetted dataset
table(rodents_subset$sex, rodents_subset$species)
#> Error: object 'rodents_subset' not found
# Original dataset
table(surveys$sex, surveys$species) # still missing a lot of rows!
#> Error: object 'surveys' not found
Created on 2025-09-11 with reprex v2.1.1
Including information about your R session
Another nice thing about {reprex} is that you can choose to include
information about your R session, in case your error has something to do
with your R settings rather than the code itself. You can do that using
the session_info argument to reprex().
For example, try running the following reprex, setting session_info = TRUE, and observe what happens.
R
library(ggplot2)
library(dplyr)
mpg %>%
ggplot(aes(x = factor(cyl), y = displ))+
geom_boxplot()
# In the console:
reprex(session_info = TRUE)2. Formatting
Two things about the formatting of the {reprex} output make it even easier to ask for help.
Displaying output
First, you’ll notice that the formatted reprex contains not only the
lines of code, but also their output. The output lines are commented out
and prefaced with a >, to differentiate them from the
code.
For example:
R
y <- 1:4
mean(y)
#> [1] 2.5
Created on 2025-09-11 with reprex v2.1.1
The whole point of a reprex is to allow the helper to run the code for themselves–if they need or want to. But when we also display the output, we make things even easier for the helper, by giving them as many tools as possible to understand our code. In some cases, the helper might even be able to diagnose the problem without running the code, just by seeing the output.
Different output formats
Second, {reprex} can provide the output in several different formats. By default, the reprex is rendered in markdown, which can easily be copied and pasted into many sites or apps. However, different places have slightly different formatting conventions for markdown. {reprex} allows you to customize the output of your reprex according to where you’re planning to post it.
The default, venue = "gh", gives you “GitHub-Flavored Markdown”,
perfect for posting on GitHub. There are other markdown “flavors” for
Slack, StackOverflow, and others.
There are some other formats, too. Specifying
venue = "r" gives you a runnable R script, with commented
output interleaved with pieces of code.
Mickey isn’t planning to post their reprex on any forums; they just
want to send it to Remy as an R script. Therefore, they render their
reprex using venue = "r".
# Mickey's reprex (1 approach)
# Required packages to run the code
library(readr)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
set.seed(1) # ensures accurate data replication
# Create a mock dataset
sample_data <- data.frame(
record_id = 1:10,
sex = sample(c('M','F', NA), 10, replace=T)
)
# The problematic code snippet
sample_subset <- sample_data %>%
filter(sex == c("F", "M"))
# Subsetted sample dataset - how many individuals for each sex?
table(sample_subset$sex)
#>
#> F
#> 1Exercise 2: Exploring reprex formatting
Prepare a simple reprex. Render it using
reprex(venue = ). Try a few different formats. You can find
more information about the different formats in the help file:
?reprex.
Callout: reprex can’t do
everything for you
People often mention {reprex} as a useful tool for creating reproducible examples, but it can’t do the work of crafting the example for you! The package doesn’t locate the problem, pare down the code, create a minimal dataset, or automatically include package dependencies.
A better way to think of {reprex} is as a tool to check your work as you go through the process of creating a reproducible example, and to help you polish up the result.
3. Context
So, we’ve established that a good minimal reproducible example should be minimal, reproducible, and formatted nicely so that helpers can easily access and read it.
But there’s one more thing to consider. Helpers won’t be thrilled if they’re presented with some code without any indication of what you’re trying to do, what went wrong, and what kind of help you need.
Let’s give them some context!
When providing context with your reprex, you should:
- Tell the helper a little bit about your problem. One sentence should be enough. What domain are you working in? What are these data about? What do the relevant variables mean?
This is particularly important if you have provided a subset of your own data instead of creating a minimal dataset from scratch. Your helper will need to interpret the column names and understand what type of data they are looking at.
Make sure to keep it minimal. Provide only the information that’s necessary to understand the problem at hand.
- Explain what you expected to happen, or what you were trying to achieve, and how it is different from what happened instead.
Remember how we diagnosed the problem in the previous episodes, identifying which lines of code showed us that something was wrong? Let’s explain to the helper what has gone wrong.
The contrast between what happened and what was supposed to happen is particularly important for semantic errors, in which the “error” is not always obvious when running the code. The code ran–but you have decided that the output is “wrong” somehow, or that it “didn’t work”. Why? How do you know that? Your helper needs to know that what you got was not what you expected, and they need to know what you expected in order to help you achieve that outcome.
Here’s an example where context is key. Imagine you’re a helper, and
someone sends you the following reprex, which includes the code to make
a boxplot from the mpg dataset.
R
library(ggplot2)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
mpg %>%
ggplot(aes(x = class, y = displ, color = class))+
geom_boxplot()

Created on 2025-09-11 with reprex v2.1.1
This example is minimal and reproducible–it’s brief, it loads all necessary packages, it uses a built-in dataset, and it is formatted nicely. But there is no obvious error! Clearly, the person asking for help has encountered a semantic error; their code runs, but it doesn’t produce what they expect. But unless they tell you how this plot differs from what they wanted, you won’t be able to help.
You ask for further context, and eventually the person seeking help replies: “I want to make a boxplot where each of the categories has its own color. But even though I set color = class here, only the outlines of the boxplots got colored in, and the inside is still white. How do I change this so that the whole box is colored in?”
Now that you know what the problem is, you can start trying to help.
Exercise 3: What makes a good description?
For each of the following reprexes, improve the description given.
“I’m trying to plot the displacements of different cars. I made this boxplot, but the boxes are showing up in the wrong order. How do I fix this? Here is my minimal reproducible example.”
R
library(ggplot2)
library(dplyr)
mpg %>%
ggplot(aes(x = class, y = displ, color = class))+
geom_boxplot()
“I’m working with this data about cars. The class column
refers to the type of car–for example,”compact” class means that the car
is quite small, while “pickup” would be a pickup truck. For each car, I
also have information about the city and highway mileage, and the
transmission, and the number of cylinders, as well as the displacement.
This dataset has 234 rows and 11 columns, although this is an example
dataset because my real dataset is much larger and has more like 500,000
rows. Anyway, in this example, I want to make a boxplot of displacement
where each of the categories has its own color. But even though I set
color = class here, only the outlines of the boxplots got colored in,
and the inside is still white. How do I make the inside a different
color? Here’s a reprex.”
R
library(ggplot2)
library(dplyr)
mpg %>%
ggplot(aes(x = class, y = displ, color = class))+
geom_boxplot()
“Help, my code isn’t working! It says I have too many elements. I made a reprex so you can see the data and the error message. I hope that’s helpful. Thank you so much!”
R
library(ggplot2)
table(mpg)
ERROR
Error in table(mpg): attempt to make a table with >= 2^31 elementsLet’s help Mickey add some context to their reprex so they’ll be ready to send it to Remy.
Exercise 4: Adding context
Working with the person next to you, write a brief description of Mickey’s problem that they could include with their reprex when they post it online.
Make sure that the description gives a little bit of background, describes what Mickey was trying to achieve, and describes what happened instead.
When you’re done, compare notes between the groups and see if you can come up with a final reprex for Mickey!
For the following exercise, you will need to choose a method for your learners to practice posting their reprexes. You should set this up before doing this exercise.
Some ideas:
Create a discussion or an issue on Github and have learners post their reprex as a response. This will require that they make a Github account.
If you have a Slack channel for your course or workshop, have learners post them there. Alternatively, post to one of the chat channels of the Data Science Learning Community. Make sure to have students respond to a thread you create, and clarify your purpose in the first post.
Create a thread on Posit Community; learners respond to the thread. See this one for example. Please be respectful–clarify the thread’s purpose at the beginning, tag it as “teaching” and “reprex-practice” or similar, and ask that others not response. Requires learners to sign up for Posit Community (free).
Have learners create rtf or R script formatted reprexes and email them to each other, then try to run each other’s reprexes in their own RStudio sessions.
We do NOT recommend posting reprexes on StackOverflow for the purpose of this exercise–StackOverflow seeks to minimize duplicate posts and would not welcome a thread like this.
Exercise 5: Putting it all together
Create a reprex (using Mickey’s example or your own brief idea). Post
it, following your instructor’s directions. Be sure to provide
appropriate context and use the appropriate venue
argument.
- The
reprexpackage makes it easy to format and share your reproducible examples. - The
reprexpackage helps you test whether your reprex is reproducible, and also helps you prepare the reprex to share with others. - Following a certain set of steps will make your questions clearer and likelier to get answered.
