Channels
Last updated on 2024-08-23 | Edit this page
Overview
Questions
- How do I move data around in Nextflow?
- How do I handle different types of input, e.g. files and parameters?
- How do I create a Nextflow channel?
- How can I use pattern matching to select input files?
- How do I change the way inputs are handled?
Objectives
- Understand how Nextflow manages data using channels.
- Understand the different types of Nextflow channels.
- Create a value and queue channel using channel factory methods.
- Select files as input based on a glob pattern.
- Edit channel factory arguments to alter how data is read in.
Channels
Earlier we learnt that channels are the way in which Nextflow sends
data around a workflow. Channels connect processes via their inputs and
outputs. Channels can store multiple items, such as files (e.g., fastq
files) or values. The number of items a channel stores determines how
many times a process will run using that channel as input.
Note: When the process runs using one item from the input
channel, we will call that run a task.
Why use Channels?
Channels are how Nextflow handles file management, allowing complex tasks to be split up, run in parallel, and reduces ‘admin’ required to get the right inputs to the right parts of the pipeline.
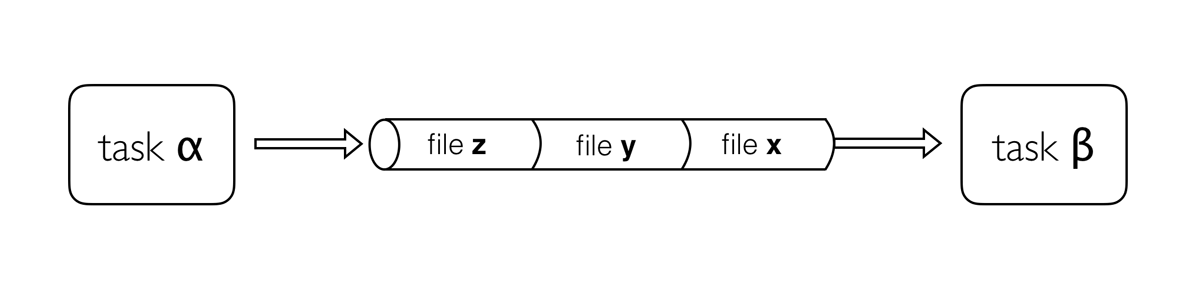
Channels are asynchronous, which means that outputs from a set of processes will not necessarily be produced in the same order as the corresponding inputs went in. However, the first element into a channel queue is the first out of the queue (First in - First out). This allows processes to run as soon as they receive input from a channel. Channels only send data in one direction, from a producer (a process/operator), to a consumer (another process/operator).
Channel types
Nextflow distinguishes between two different kinds of channels: queue channels and value channels.
Queue channel
Queue channels are a type of channel in which data is consumed (used up) to make input for a process/operator. Queue channels can be created in two ways:
- As the outputs of a process.
- Explicitly using channel factory methods such as Channel.of or Channel.fromPath.
Value channels
The second type of Nextflow channel is a value channel.
A value channel is bound to a single
value. A value channel can be used an unlimited number times since its
content is not consumed. This is also useful for processes that need to
reuse input from a channel, for example, a reference genome sequence
file that is required by multiple steps within a process, or by more
than one process.
Queue vs Value Channel.
What type of channel would you use to store the following?
- Multiple values.
- A list with one or more values.
- A single value.
- A queue channels is used to store multiple values.
- A value channel is used to store a single value, this can be a list with multiple values.
- A value channel is used to store a single value.
Creating Channels using Channel factories
Channel factories are used to explicitly create channels. In
programming, factory methods (functions) are a programming design
pattern used to create different types of objects (in this case,
different types of channels). They are implemented for things that
represent more generalised concepts, such as a Channel.
Channel factories are called using the
Channel.<method> syntax, and return a specific
instance of a Channel.
The value Channel factory
The value factory method is used to create a value
channel. Values are put inside parentheses () to assign
them to a channel.
For example:
GROOVY
ch1 = Channel.value( 'GRCh38' )
ch2 = Channel.value( ['chr1', 'chr2', 'chr3', 'chr4', 'chr5'] )
ch3 = Channel.value( ['chr1' : 248956422, 'chr2' : 242193529, 'chr3' : 198295559] )- Creates a value channel and binds a string to it.
- Creates a value channel and binds a list object to it that will be emitted as a single item.
- Creates a value channel and binds a map object to it that will be emitted as a single item.
The value method can only take 1 argument, however, this can be a single list or map containing several elements.
Reminder:
- A List
object can be defined by placing the values in square brackets
[]separated by a comma. - A Map
object is similar, but with
key:value pairsseparated by commas.
To view the contents of a value channel, use the view
operator. We will learn more about channel operators in a later
episode.
GROOVY
ch1 = Channel.value( 'GRCh38' )
ch2 = Channel.value( ['chr1', 'chr2', 'chr3', 'chr4', 'chr5'] )
ch3 = Channel.value( ['chr1' : 248956422, 'chr2' : 242193529, 'chr3' : 198295559] )
ch1.view()
ch2.view()
ch3.view()Each item in the channel is printed on a separate line.
OUTPUT
GRCh38
[chr1, chr2, chr3, chr4, chr5]
[chr1:248956422, chr2:242193529, chr3:198295559]Queue channel factory
Queue (consumable) channels can be created using the following channel factory methods.
Channel.ofChannel.fromListChannel.fromPathChannel.fromFilePairsChannel.fromSRA
The of Channel factory
When you want to create a channel containing multiple values you can
use the channel factory Channel.of. Channel.of
allows the creation of a queue channel with the values
specified as arguments, separated by a ,.
OUTPUT
chr1
chr3
chr5
chr7The first line in this example creates a variable
chromosome_ch. chromosome_ch is a queue
channel containing the four values specified as arguments in the
of method. The view operator will print one
line per item in a list. Therefore the view operator on the
second line will print four lines, one for each element in the
channel:
You can specify a range of numbers as a single argument using the
Groovy range operator ... This creates each value in the
range (including the start and end values) as a value in the channel.
The Groovy range operator can also produce ranges of dates, letters, or
time. More information on the range operator can be found here.
Arguments passed to the of method can be of varying
types e.g., combinations of numbers, strings, or objects. In the above
examples we have examples of both string and number data types.
Channel.from
You may see the method Channel.from in older nextflow
scripts. This performs a similar function but is now deprecated (no
longer used), and so Channel.of should be used instead.
Create a value and Queue and view Channel contents
- Create a Nextflow script file called
channel.nf. - Create a Value channel
ch_vlcontaining the String'GRCh38'. - Create a Queue channel
ch_qucontaining the values 1 to 4. - Use
.view()operator on the channel objects to view the contents of the channels. - Run the code using
The fromList Channel factory
You can use the Channel.fromList method to create a
queue channel from a list object.
GROOVY
aligner_list = ['salmon', 'kallisto']
aligner_ch = Channel.fromList(aligner_list)
aligner_ch.view()This would produce two lines.
OUTPUT
salmon
kallistoChannel.fromList vs Channel.of
In the above example, the channel has two elements. If you has used
the Channel.of(aligner_list) it would have contained only 1 element
[salmon, kallisto] and any operator or process using the
channel would run once.
Creating channels from a list
Write a Nextflow script that creates both a queue and
value channel for the list
Then print the contents of the channels using the view
operator. How many lines does the queue and value channel print?
Hint: Use the fromList() and
value() Channel factory methods.
GROOVY
ids = ['ERR908507', 'ERR908506', 'ERR908505']
queue_ch = Channel.fromList(ids)
value_ch = Channel.value(ids)
queue_ch.view()
value_ch.view()OUTPUT
N E X T F L O W ~ version 21.04.0
Launching `channel_fromList.nf` [wise_hodgkin] - revision: 22d76be151
ERR908507
ERR908506
ERR908505
[ERR908507, ERR908506, ERR908505]The queue channel queue_ch will print three lines.
The value channel value_ch will print one line.
The fromPath Channel factory
The previous channel factory methods dealt with sending general
values in a channel. A special channel factory method
fromPath is used when wanting to pass files.
The fromPath factory method creates a queue
channel containing one or more files matching a file path.
The file path (written as a quoted string) can be the location of a single file or a “glob pattern” that matches multiple files or directories.
The file path can be a relative path (path to the file from the
current directory), or an absolute path (path to the file from the
system root directory - starts with /).
The script below creates a queue channel with a single file as its content.
OUTPUT
data/yeast/reads/ref1_2.fq.gzYou can also use glob syntax to specify pattern-matching behaviour for files. A glob pattern is specified as a string and is matched against directory or file names.
- An asterisk,
*, matches any number of characters (including none). - Two asterisks,
**, works like * but will also search sub directories. This syntax is generally used for matching complete paths. - Braces
{}specify a collection of subpatterns. For example:{bam,bai}matches “bam” or “bai”
For example the script below uses the *.fq.gz pattern to
create a queue channel that contains as many items as there are files
with .fq.gz extension in the data/yeast/reads
folder.
OUTPUT
data/yeast/reads/ref1_2.fq.gz
data/yeast/reads/etoh60_3_2.fq.gz
data/yeast/reads/temp33_1_2.fq.gz
data/yeast/reads/temp33_2_1.fq.gz
data/yeast/reads/ref2_1.fq.gz
data/yeast/reads/temp33_3_1.fq.gz
[..truncated..]Note The pattern must contain at least a star wildcard character.
You can change the behaviour of Channel.fromPath method
by changing its options. A list of .fromPath options is
shown below.
Available fromPath options:
| Name | Description |
|---|---|
| glob | When true, the characters *, ?,
[] and {} are interpreted as glob wildcards,
otherwise they are treated as literal characters (default: true) |
| type | The type of file paths matched by the string, either
file, dir or any (default:
file) |
| hidden | When true, hidden files are included in the resulting paths (default: false) |
| maxDepth | Maximum number of directory levels to visit (default: no limit) |
| followLinks | When true, symbolic links are followed during directory tree traversal, otherwise they are managed as files (default: true) |
| relative | When true returned paths are relative to the top-most common directory (default: false) |
| checkIfExists | When true throws an exception if the specified path does not exist in the file system (default: false) |
We can change the default options for the fromPath
method to give an error if the file doesn’t exist using the
checkIfExists parameter. In Nextflow, method parameters are
separated by a , and parameter values specified with a
colon :.
If we execute a Nextflow script with the contents below, it will run and not produce an output, or an error message that the file does not exist. This is likely not what we want.
OUTPUT
N E X T F L O W ~ version 20.10.0
Launching `channels.nf` [scruffy_swartz] DSL2 - revision: 2c8f18ab48Add the argument checkIfExists with the value
true.
GROOVY
read_ch = Channel.fromPath( 'data/chicken/reads/*.fq.gz', checkIfExists: true )
read_ch.view()This will give an error as there is no data/chicken directory.
OUTPUT
N E X T F L O W ~ version 20.10.0
Launching `channels.nf` [intergalactic_mcclintock] - revision: d2c138894b
No files match pattern `*.fq.gz` at path: data/chicken/reads/Using Channel.fromPath
- Create a Nextflow script
channel_fromPath.nf - Use the
Channel.fromPathmethod to create a channel containing all files in thedata/yeast/directory, including the subdirectories. - Add the parameter to include any hidden files.
- Then print all file names using the
viewoperator.
Hint: You need two asterisks, i.e. **,
to search subdirectories.
OUTPUT
N E X T F L O W ~ version 21.04.0
Launching `channel_fromPath.nf` [reverent_mclean] - revision: cf02269bcb
data/yeast/samples.csv
data/yeast/reads/etoh60_3_2.fq.gz
data/yeast/reads/temp33_1_2.fq.gz
data/yeast/reads/temp33_2_1.fq.gz
[..truncated..]The fromFilePairs Channel factory
We have seen how to process files individually using
fromPath. In Bioinformatics we often want to process files
in pairs or larger groups, such as read pairs in sequencing.
For example is the data/yeast/reads directory we have
nine groups of read pairs.
| Sample group | read1 | read2 |
|---|---|---|
| ref1 | data/yeast/reads/ref1_1.fq.gz | data/yeast/reads/ref1_2.fq.gz |
| ref2 | data/yeast/reads/ref2_1.fq.gz | data/yeast/reads/ref2_2.fq.gz |
| ref3 | data/yeast/reads/ref3_1.fq.gz | data/yeast/reads/ref3_2.fq.gz |
| temp33_1 | data/yeast/reads/temp33_1_1.fq.gz | data/yeast/reads/temp33_1_2.fq.gz |
| temp33_2 | data/yeast/reads/temp33_2_1.fq.gz | data/yeast/reads/temp33_2_2.fq.gz |
| temp33_3 | data/yeast/reads/temp33_3_1.fq.gz | data/yeast/reads/temp33_3_2.fq.gz |
| etoh60_1 | data/yeast/reads/etoh60_1_1.fq.gz | data/yeast/reads/etoh60_1_2.fq.gz |
| etoh60_2 | data/yeast/reads/etoh60_2_1.fq.gz | data/yeast/reads/etoh60_2_2.fq.gz |
| etoh60_3 | data/yeast/reads/etoh60_3_1.fq.gz | data/yeast/reads/etoh60_3_2.fq.gz |
Nextflow provides a convenient Channel factory method for this common
bioinformatics use case. The fromFilePairs method creates a
queue channel containing a tuple for every set of files
matching a specific glob pattern (e.g.,
/path/to/*_{1,2}.fq.gz).
A tuple is a grouping of data, represented as a Groovy
List.
- The first element of the tuple emitted from
fromFilePairsis a string based on the shared part of the filenames (i.e., the*part of the glob pattern). - The second element is the list of files matching the remaining part
of the glob pattern (i.e., the
<string>_{1,2}.fq.gzpattern). This will include any files ending_1.fq.gzor_2.fq.gz.
OUTPUT
[etoh60_3, [data/yeast/reads/etoh60_3_1.fq.gz, data/yeast/reads/etoh60_3_2.fq.gz]]
[temp33_1, [data/yeast/reads/temp33_1_1.fq.gz, data/yeast/reads/temp33_1_2.fq.gz]]
[ref1, [data/yeast/reads/ref1_1.fq.gz, data/yeast/reads/ref1_2.fq.gz]]
[ref2, [data/yeast/reads/ref2_1.fq.gz, data/yeast/reads/ref2_2.fq.gz]]
[temp33_2, [data/yeast/reads/temp33_2_1.fq.gz, data/yeast/reads/temp33_2_2.fq.gz]]
[ref3, [data/yeast/reads/ref3_1.fq.gz, data/yeast/reads/ref3_2.fq.gz]]
[temp33_3, [data/yeast/reads/temp33_3_1.fq.gz, data/yeast/reads/temp33_3_2.fq.gz]]
[etoh60_1, [data/yeast/reads/etoh60_1_1.fq.gz, data/yeast/reads/etoh60_1_2.fq.gz]]
[etoh60_2, [data/yeast/reads/etoh60_2_1.fq.gz, data/yeast/reads/etoh60_2_2.fq.gz]]This will produce a queue channel, read_pair_ch ,
containing nine elements.
Each element is a tuple that has;
- string value (the file prefix matched, e.g
temp33_1) - and a list with the two files e,g.
[data/yeast/reads/temp33_1_1.fq.gz, data/yeast/reads/temp33_1_2.fq.gz].
The asterisk character *, matches any number of
characters (including none), and the {} braces specify a
collection of subpatterns. Therefore the *_{1,2}.fq.gz
pattern matches any file name ending in _1.fq.gz or
_2.fq.gz .
What if you want to capture more than a pair?
If you want to capture more than two files for a pattern you will
need to change the default size argument (the default value
is 2) to the number of expected matching files.
For example in the directory data/yeast/reads there are
six files with the prefix ref. If we want to group (create
a tuple) for all of these files we could write;
GROOVY
read_group_ch = Channel.fromFilePairs('data/yeast/reads/ref{1,2,3}*',size:6)
read_group_ch.view()The code above will create a queue channel containing one element. The element is a tuple of which contains a string value, that is the pattern ref, and a list of six files matching the pattern.
OUTPUT
[ref, [data/yeast/reads/ref1_1.fq.gz, data/yeast/reads/ref1_2.fq.gz, data/yeast/reads/ref2_1.fq.gz, data/yeast/reads/ref2_2.fq.gz, data/yeast/reads/ref3_1.fq.gz, data/yeast/reads/ref3_2.fq.gz]]See more information about the channel factory
fromFilePairs here
More complex patterns
If you need to match more complex patterns you should create a sample sheet specifying the files and create a channel from that. This will be covered in the operator episode.
Create a channel containing groups of files
- Create a Nextflow script file
channel_fromFilePairs.nf. - Use the
fromFilePairsmethod to create a channel containing three tuples. Each tuple will contain the pairs of fastq reads for the three temp33 samples in thedata/yeast/readsdirectory
OUTPUT
N E X T F L O W ~ version 21.04.0
Launching `channels.nf` [stupefied_lumiere] - revision: a3741edde2
[temp33_1, [data/yeast/reads/temp33_1_1.fq.gz, data/yeast/reads/temp33_1_2.fq.gz]]
[temp33_3, [data/yeast/reads/temp33_3_1.fq.gz, data/yeast/reads/temp33_3_2.fq.gz]]
[temp33_2, [data/yeast/reads/temp33_2_1.fq.gz, data/yeast/reads/temp33_2_2.fq.gz]]Key Points
- Channels must be used to import data into Nextflow.
- Nextflow has two different kinds of channels: queue channels and value channels.
- Data in value channels can be used multiple times in workflow.
- Data in queue channels are consumed when they are used by a process or an operator.
- Channel factory methods, such as
Channel.of, are used to create channels. - Channel factory methods have optional parameters e.g.,
checkIfExists, that can be used to alter the creation and behaviour of a channel.
