Plotting
Overview
Teaching: 25 min
Exercises: 15 minQuestions
How do I create plots in Python?
Objectives
Create basic plots in Python using the Matplotlib library
There are several different Python plotting libraries. Perhaps the most popular one is Matplotlib, initially released back in 2003. Another library, called Seaborn, is based on Matplotlib, and provides “nicer” defaults for colors and plots, making it easier to build beautiful publication-ready plots. A modern alternative is Plotly, that is available not only in Python, but also in R, Julia, JavaScript, MatLab, and F#. This tutorial serves as a brief introduction into Matplotlib.
Introduction to plots: histograms
We’ll use a dataset of reference genomes of Streptomyces. To download it and load it into the Python environment, we can use the Pandas library.
import pandas as pd
data = pd.read_csv("https://raw.githubusercontent.com/carpentries-incubator/pangenomics-python/gh-pages/data/streptomyces.csv")
data
Matplotlib is a large library; however, most plotting functions are available
in the matplotlib.pyplot module, which is usually imported as follows.
import matplotlib.pyplot as plt
The Matplotlib official website provides a convenient page
showing the main plot types available. We’ll begin with a histogram showing the
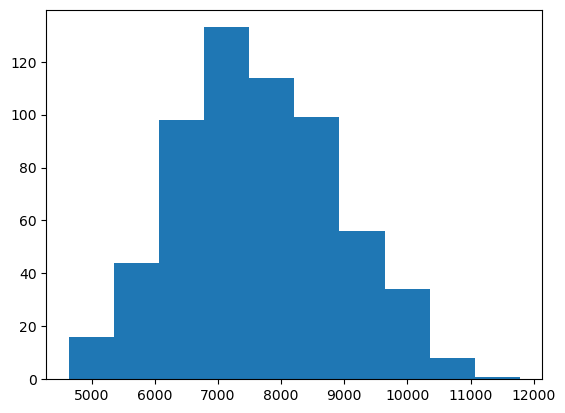
number of genes per reference assembly, which can be accomplished by using the
plt.hist function and pass the "genes" column of the dataset, grouping the
values automatically. Use the plt.show() function to draw the plot on the
screen.
plt.hist(data["genes"])
plt.show()
The plt.hist function has many many options
that allow you to customize how the chart looks like. For instance, we can use
the bins parameter to set the number of bars in the histogram. Furthermore,
plots are useless without proper labels, so we’ll use the plt.xlabel,
plt.ylabel and plt.title functions to define the label for the x axis, the
label for the y axis, and the title for the plot, respectively.
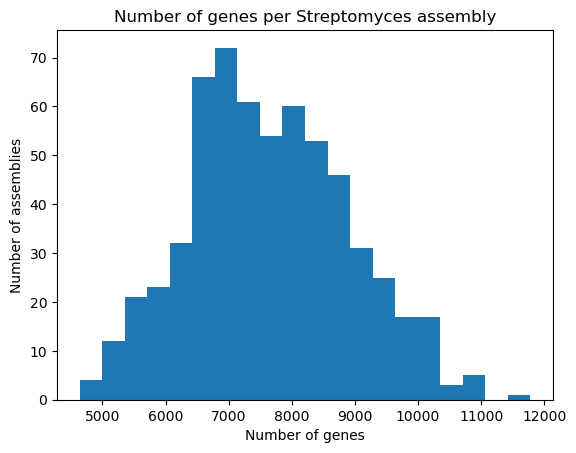
plt.hist(data["genes"], bins=20)
plt.xlabel("Number of genes")
plt.ylabel("Number of assemblies")
plt.title("Number of genes per Streptomyces assembly")
plt.show()
Bars and lines
Whereas plt.hist allows you to pass the variables directly, other plot types
require you to perform some manipulations on the dataset, because we should
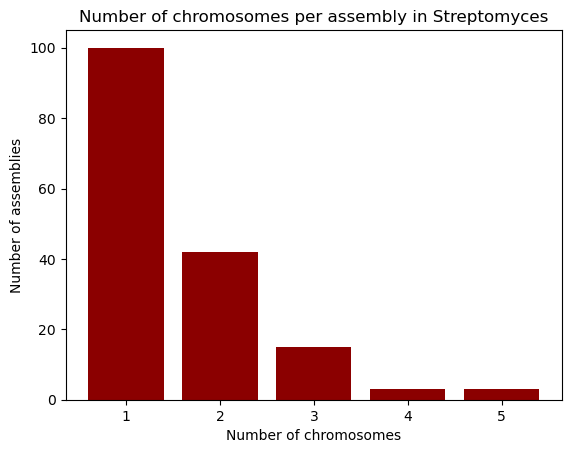
explicitly provide both the x and y axes. The plt.bar function, as its name
suggests, creates bar plots; we’ll use it to visualize the number of chromosomes
per assembly in our dataset. First, we take the "chromosomes" column from the
dataset, and use the .value_counts() method to count how many times each
chromosome count appears in it. This method returns a Pandas Series, with an
index and values which we can access. So, in order to build the bar plot,
we first provide the unique chromosome counts for the x axis, and the values for
the y axis. We’ll also change the bar colors to dark red and add labels.
chromosomes = data["chromosomes"].value_counts()
chromosomes
chromosomes
1.0 100
2.0 42
3.0 15
4.0 3
5.0 3
Name: count, dtype: int64
plt.bar(chromosomes.index, chromosomes.values, color="darkred")
plt.xlabel("Number of chromosomes")
plt.ylabel("Number of assemblies")
plt.title("Number of chromosomes per assembly in Streptomyces")
plt.show()
Going horizontal: creating horizontal bar plots
If you wish to use a horizontal bar plot instead of a vertical one, use the
plt.barhfunction. Don’t forget to change your labels accordingly!
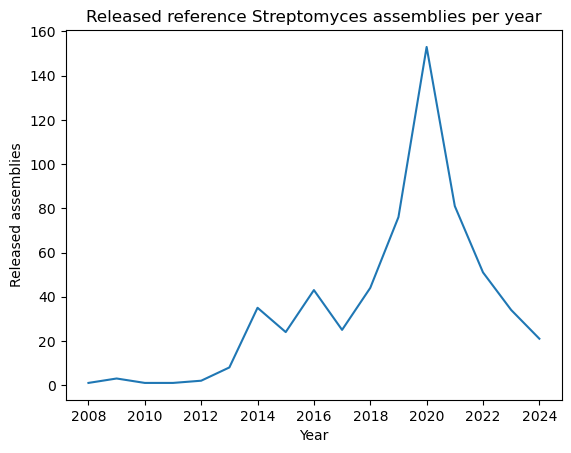
Let’s now learn how to build line plots using plt.plot
by visualizing the number of reference assemblies released by year. Similar to
the previous example, we’ll use the .value_counts method on the
"release_year" columns to count the number of assemblies per year; however,
this method sorts the index by the count, so in order to keep the original order
(which is already chronological), we pass the sort=False parameter. Next,
we provide the index and values for the x and y axes of our plot.
genomes_year = data["release_year"].value_counts(sort=False)
genomes_year
release_year
2008 1
2009 3
2010 1
2011 1
2012 2
2013 8
2014 35
2015 24
2016 43
2017 25
2018 44
2019 76
2020 153
2021 81
2022 51
2023 34
2024 21
Name: count, dtype: int64
plt.plot(genomes_year.index, genomes_year.values)
plt.xlabel("Year")
plt.ylabel("Released assemblies")
plt.title("Released reference Streptomyces assemblies per year")
plt.show()
A nice feature about plt.plot is that we can change the way the line looks
like, either by modifying the edges and/or the vertices. You can find the format
guide in the “Format Strings” section of the function’s documentation. Some example
strings you can use are depicted in the next code block.
"--" # Dashed line
":" # Dotted line
"o" # Large dots only
"v" # Down-facing triangles only
"s" # Squares only
"--o" # Dashed line with large dots
":s" # Dotted line with squares
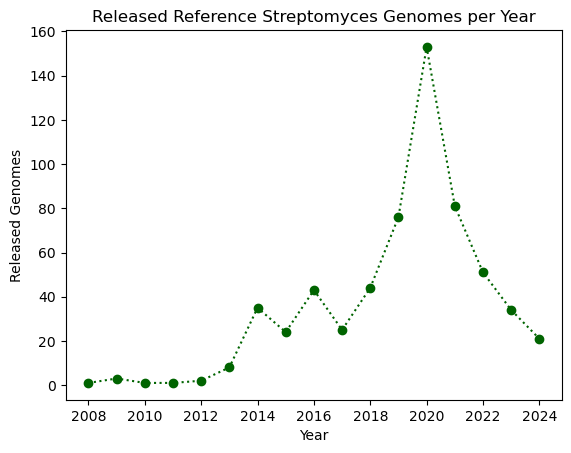
Let’s modify our plot by making it dotted with large dots, with a dark green color.
plt.plot(genomes_year.index, genomes_year.values, ':o', color="darkgreen")
plt.xlabel("Year")
plt.ylabel("Released Genomes")
plt.title("Released Reference Streptomyces Genomes per Year")
plt.show()
Exercise (Beginner): Plotting with Matplotlib
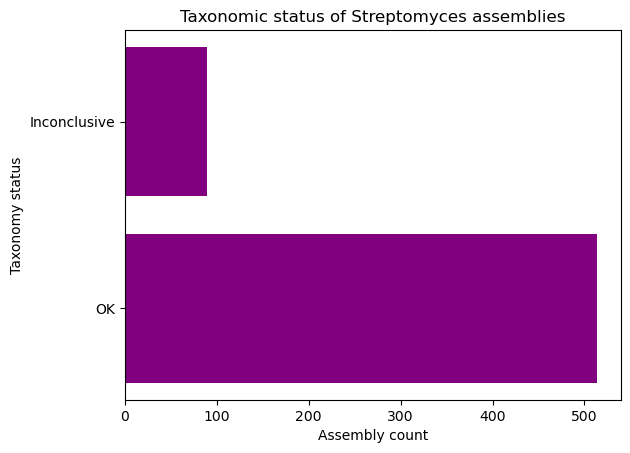
Complete the following code block to create a horizontal bar plot with the number of assemblies with conclusive and inconclusive taxonomy from the dataset. Use purple to color the bars.
taxonomy = data["taxonomy_status"].________() plt.________(taxonomy.________, taxonomy.__________, ________="purple") plt.________("Taxonomy status") plt.________("Assembly count") plt.title("Taxonomic status of Streptomyces assemblies") plt.show()Solution
taxonomy = data["taxonomy_status"].value_counts() plt.barh(taxonomy.index, taxonomy.values, color="purple") plt.ylabel("Taxonomy status") plt.xlabel("Assembly count") plt.title("Taxonomic status of Streptomyces assemblies") plt.show()
Multiple plots in a single figure
Extra content
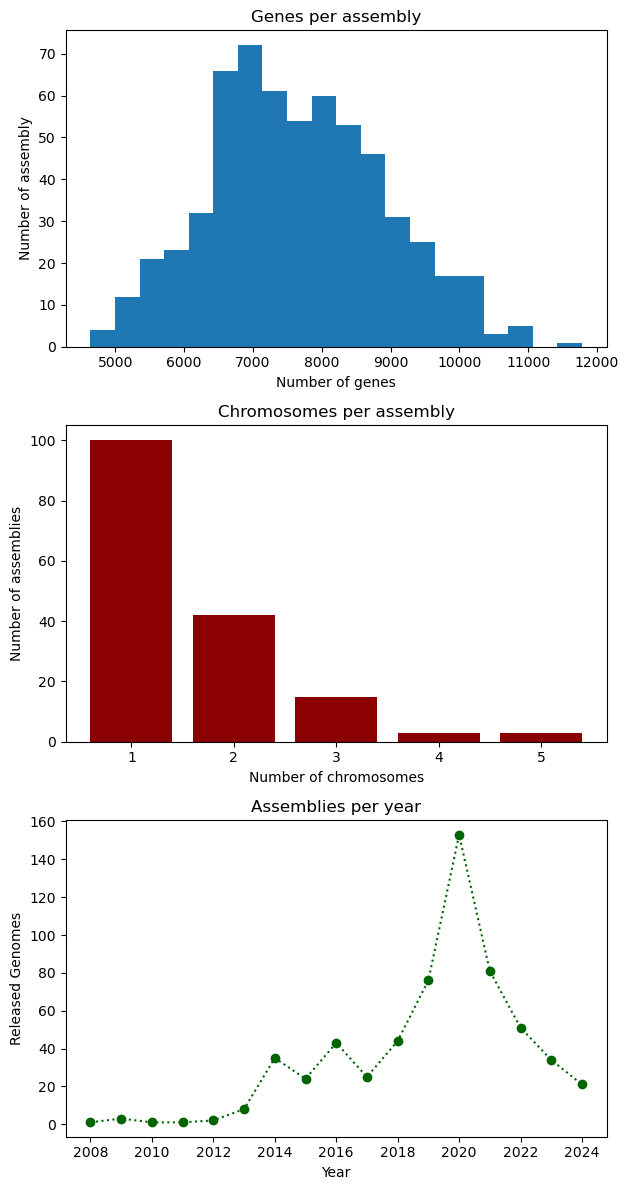
The
plt.subplots(x, y)function creates a multi-plot figure withxrows andycolumns. It returns aFigureobject that allows to modify general aspects of the figure, and an empty array which will store the plots and are accessible via indices. As an example, we’ll create a figure with one column and three rows and place the three plots be made in the lesson. Instead of using.xlabel,.ylabeland.title, we use.set_xlabel,.set_ylabeland.set_title, respectively. At the end, we use theset_figheightmethod to set the height for the entire figure, and the.tight_layoutmethod on the figure in order to ensure that everything fits in properly.# Figure initialization fig, ax = plt.subplots(3) # First plot: histogram ax[0].hist(data["genes"], bins=20) ax[0].set_xlabel("Number of genes") ax[0].set_ylabel("Number of assembly") ax[0].set_title("Genes per assembly") # Second plot: bar ax[1].bar(chromosomes.index, chromosomes.values, color="darkred") ax[1].set_xlabel("Number of chromosomes") ax[1].set_ylabel("Number of assemblies") ax[1].set_title("Chromosomes per assembly") # Third plot: line ax[2].plot(genomes_year.index, genomes_year.values, ':o', color="darkgreen") ax[2].set_xlabel("Year") ax[2].set_ylabel("Released Genomes") ax[2].set_title("Assemblies per year") # Figure configuration fig.set_figheight(12) fig.tight_layout() plt.show()
Key Points
Matplotlib is a popular plotting library for Python