1.1 Introduction to Our Software Project
Last updated on 2025-07-30 | Edit this page
Estimated time: 30 minutes
Overview
Questions
- What is the design architecture of our example software project?
- Why is splitting code into smaller functional units (modules) good when designing software?
Objectives
- Use Git to obtain a working copy of our software project from GitHub.
- Inspect the structure and architecture of our software project.
- Understand Model-View-Controller (MVC) architecture in software design and its use in our project.
Patient Inflammation Study Project
You have joined a software development team that has been working on the patient inflammation study project developed in Python and stored on GitHub. The project analyses the data to study the effect of a new treatment for arthritis by analysing the inflammation levels in patients who have been given this treatment. It reuses the inflammation datasets from the Software Carpentry Python novice lesson.
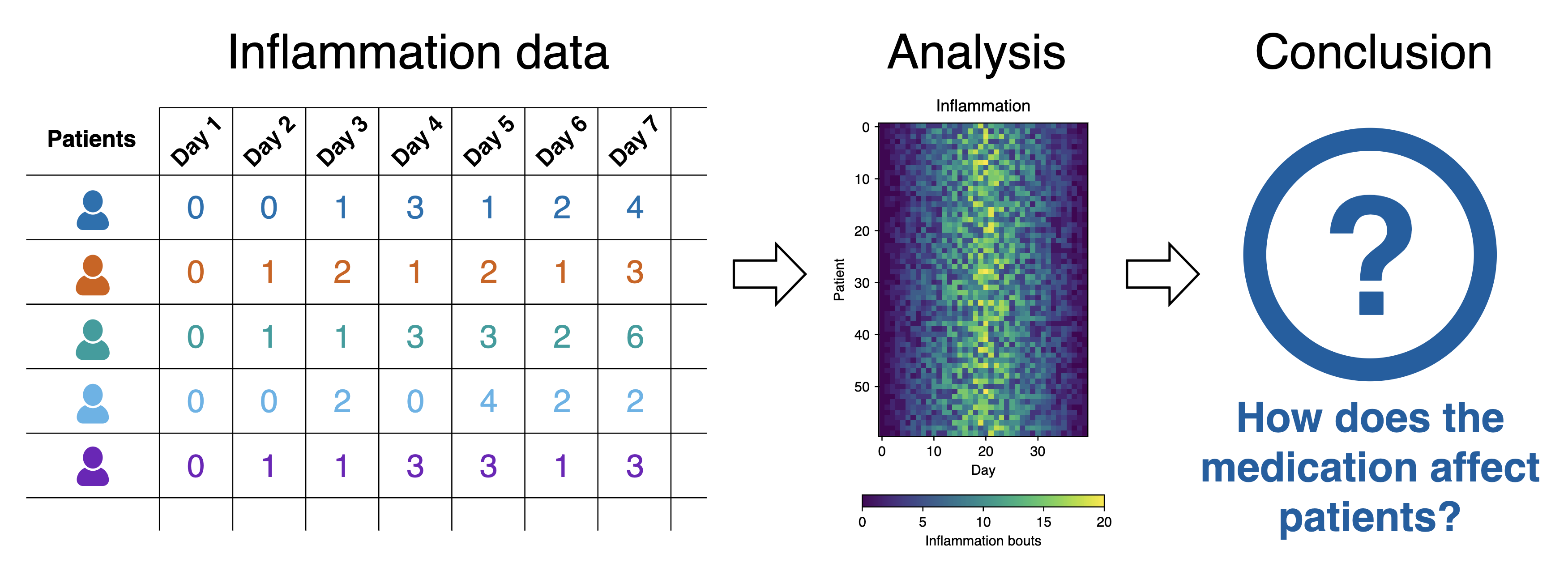
Inflammation study pipeline from the Software Carpentry Python novice lesson
What Does Patient Inflammation Data Contain?
Each dataset records inflammation measurements from a separate clinical trial of the drug, and each dataset contains information for 60 patients, who had their inflammation levels recorded (in some arbitrary units of inflammation measurement) for 40 days whilst participating in the trial. A snapshot of one of the data files is shown in the diagram above.
Each of the data files uses the popular comma-separated (CSV) format to represent the data, where:
- each row holds inflammation measurements for a single patient
- each column represents a successive day in the trial
- each cell represents an inflammation reading on a given day for a patient
The project is not finished and contains some errors. You will be working on your own and in collaboration with others to fix and build on top of the existing code during the course.
Downloading Our Software Project
To start working on the project, you will first create a fork of the software project repository from GitHub within your own GitHub account and then obtain a local copy of that project (from your GitHub) on your machine.
- Make sure you have a GitHub account and that you have set up SSH key pair for authentication with GitHub.
Note: while it is possible to use HTTPS with a personal access token for authentication with GitHub, the recommended and supported authentication method to use for this course is SSH with key pairs.
Log into your GitHub account.
Go to the software project repository in GitHub.
- Click the
Forkbutton towards the top right of the repository’s GitHub page to create a fork of the repository under your GitHub account. Remember, you will need to be signed into GitHub for theForkbutton to work.
Note: each participant is creating their own fork of the project to work on.
Note 2: we are creating a fork of the software project repository (instead of copying it from its template) because we want to preserve the history of all commits (with template copying you only get a snapshot of a repository at a given point in time).
- Make sure to select your personal account and set the name of the
project to
python-intermediate-inflammation(you can call it anything you like, but it may be easier for future group exercises if everyone uses the same name). Ensure that you uncheck theCopy the main branch onlyoption. This guarantees you get all the branches from this repository needed for later exercises.
Click the
Create forkbutton and wait for GitHub to create the forked copy of the repository under your account.Locate the forked repository under your own GitHub account. GitHub should redirect you there automatically after creating the fork. If this does not happen, click your user icon in the top right corner and select
Your Repositoriesfrom the drop-down menu, then locate your newly created fork.
Exercise: Obtain the Software Project Locally
Using the command line, clone the copied repository from your GitHub account into the home directory on your computer using SSH. Which command(s) would you use to get a detailed list of contents of the directory you have just cloned?
- Find the SSH URL of the software project repository to clone from your GitHub account. Make sure you do not clone the original repository but rather your own fork, as you should be able to push commits to it later on. Also make sure you select the SSH tab and not the HTTPS one. For this course, SSH is the preferred way of authenticating when sending your changes back to GitHub. If you have only authenticated through HTTPS in the past, please follow the guidance at the top of this section to add an SSH key to your GitHub account.
- Make sure you are located in your home directory in the command line with:
- From your home directory in the command line, do:
Make sure you are cloning your fork of the software project and not the original repository.
- Navigate into the cloned repository folder in your command line with:
Note: If you have accidentally copied the HTTPS URL of your repository instead of the SSH one, you can easily fix that from your project folder in the command line with:
Our Software Project’s Structure
Let’s inspect the content of the software project from the command
line. From the root directory of the project, you can use the command
ls -l to get a more detailed list of the contents. You
should see something similar to the following.
BASH
$ cd ~/python-intermediate-inflammation
$ ls -l
total 24
-rw-r--r-- 1 carpentry users 1055 20 Apr 15:41 README.md
drwxr-xr-x 18 carpentry users 576 20 Apr 15:41 data
drwxr-xr-x 5 carpentry users 160 20 Apr 15:41 inflammation
-rw-r--r-- 1 carpentry users 1122 20 Apr 15:41 inflammation-analysis.py
drwxr-xr-x 4 carpentry users 128 20 Apr 15:41 testsAs can be seen from the above, our software project contains the
README file (that typically describes the project, its
usage, installation, authors and how to contribute), Python script
inflammation-analysis.py, and three directories -
inflammation, data and tests.
The Python script inflammation-analysis.py provides the
main entry point in the application, and on closer inspection, we can
see that the inflammation directory contains two more
Python scripts - views.py and models.py. We
will have a more detailed look into these shortly.
BASH
$ ls -l inflammation
total 24
-rw-r--r-- 1 alex staff 71 29 Jun 09:59 __init__.py
-rw-r--r-- 1 alex staff 838 29 Jun 09:59 models.py
-rw-r--r-- 1 alex staff 649 25 Jun 13:13 views.pyDirectory data contains several files with patients’
daily inflammation information (along with some other files):
BASH
$ ls -l data
total 264
-rw-r--r-- 1 alex staff 5365 25 Jun 13:13 inflammation-01.csv
-rw-r--r-- 1 alex staff 5314 25 Jun 13:13 inflammation-02.csv
-rw-r--r-- 1 alex staff 5127 25 Jun 13:13 inflammation-03.csv
-rw-r--r-- 1 alex staff 5367 25 Jun 13:13 inflammation-04.csv
-rw-r--r-- 1 alex staff 5345 25 Jun 13:13 inflammation-05.csv
-rw-r--r-- 1 alex staff 5330 25 Jun 13:13 inflammation-06.csv
-rw-r--r-- 1 alex staff 5342 25 Jun 13:13 inflammation-07.csv
-rw-r--r-- 1 alex staff 5127 25 Jun 13:13 inflammation-08.csv
-rw-r--r-- 1 alex staff 5327 25 Jun 13:13 inflammation-09.csv
-rw-r--r-- 1 alex staff 5342 25 Jun 13:13 inflammation-10.csv
-rw-r--r-- 1 alex staff 5127 25 Jun 13:13 inflammation-11.csv
-rw-r--r-- 1 alex staff 5340 25 Jun 13:13 inflammation-12.csv
-rw-r--r-- 1 alex staff 22554 25 Jun 13:13 python-novice-inflammation-data.zip
-rw-r--r-- 1 alex staff 12 25 Jun 13:13 small-01.csv
-rw-r--r-- 1 alex staff 15 25 Jun 13:13 small-02.csv
-rw-r--r-- 1 alex staff 12 25 Jun 13:13 small-03.csvAs previously mentioned, each of the inflammation data files contains separate trial data for 60 patients over 40 days.
Exercise: Have a Peek at the Data
Which command(s) would you use to list the contents or a first few
lines of data/inflammation-01.csv file?
- To list the entire content of a file from the project root do:
cat data/inflammation-01.csv. - To list the first 5 lines of a file from the project root do:
OUTPUT
0,0,1,3,1,2,4,7,8,3,3,3,10,5,7,4,7,7,12,18,6,13,11,11,7,7,4,6,8,8,4,4,5,7,3,4,2,3,0,0
0,1,2,1,2,1,3,2,2,6,10,11,5,9,4,4,7,16,8,6,18,4,12,5,12,7,11,5,11,3,3,5,4,4,5,5,1,1,0,1
0,1,1,3,3,2,6,2,5,9,5,7,4,5,4,15,5,11,9,10,19,14,12,17,7,12,11,7,4,2,10,5,4,2,2,3,2,2,1,1
0,0,2,0,4,2,2,1,6,7,10,7,9,13,8,8,15,10,10,7,17,4,4,7,6,15,6,4,9,11,3,5,6,3,3,4,2,3,2,1
0,1,1,3,3,1,3,5,2,4,4,7,6,5,3,10,8,10,6,17,9,14,9,7,13,9,12,6,7,7,9,6,3,2,2,4,2,0,1,1Directory tests contains several tests that have been
implemented already. We will be adding more tests during the course as
our code grows.
BASH
$ ls -l tests
total 16
-rw-r--r-- 1 alex staff 941 18 Dec 11:42 test_models.py
-rw-r--r-- 1 alex staff 182 18 Dec 11:42 test_patient.pyAn important thing to note here is that the structure of our project is not arbitrary. One of the big differences between novice and intermediate software development is planning the structure of your code. This structure includes software components and behavioural interactions between them, including how these components are laid out in a directory and file structure. A novice will often make up the structure of their code as they go along. However, for more advanced software development, we need to plan and design this structure - called a software architecture - beforehand.
Let us have a quick look into what a software architecture is and which architecture is used by our software project before we start adding more code to it.
Software Architecture
A software architecture is the fundamental structure of a software system that is decided at the beginning of project development based on its requirements and cannot be changed that easily once implemented. It refers to a “bigger picture” of a software system that describes high-level components (modules) of the system and how they interact.
In software design and development, large systems or programs are
often decomposed into a set of smaller modules each with a subset of
functionality. Typical examples of modules in programming are software
libraries; some software libraries, such as numpy and
matplotlib in Python, are bigger modules that contain
several smaller sub-modules. Another example of modules are classes in
object-oriented programming languages.
Programming Modules and Interfaces
Although modules are self-contained and independent elements to a large extent (they can depend on other modules), there are well-defined ways of how they interact with one another. These rules of interaction are called programming interfaces - they define how other modules (clients) can use a particular module. Typically, an interface to a module includes rules on how a module can take input from and how it gives output back to its clients. A client can be a human, in which case we also call these user interfaces. Even smaller functional units such as functions/methods have clearly defined interfaces - a function/method’s definition (also known as a signature) states what parameters it can take as input and what it returns as an output.
We are going to talk about software architecture and design a bit more in Section 3 - for now it is sufficient to know that the way our software project’s code is structured is intentional.
Our Project’s Architecture
Our software project uses the Model-View-Controller (MVC) architecture. MVC architecture divides the software logic into three interconnected modules:
- Model (data) - represents the data used by a program and contains operations/rules for manipulating and changing the data in the model (a database, a file, a single data object or a series of objects - for example a table representing patients’ data).
- View (client interface) - provides means of displaying data to users/clients within an application (i.e. provides visualisation of the state of the model). For example, displaying a window with input fields and buttons (Graphical User Interface, GUI) or textual options within a command line (Command Line Interface, CLI) are examples of Views.
- Controller (processes that handle input/output and manipulate the data) - accepts input from the View and performs the corresponding action on the Model (changing the state of the model) and then updates the View accordingly.
In our project, inflammation-analysis.py is the
Controller module that performs basic statistical
analysis over patient data and provides the main entry point into the
application. The View and Model
modules are contained in the files views.py and
models.py, respectively, and are conveniently named. Data
underlying the Model is contained within the directory
data - as we have seen already it contains several files
with patients’ daily inflammation information.
We will revisit the software architecture and MVC topics once again in later episodes when we talk in more detail about software architecture and design. We now proceed to set up our virtual development environment and start working with the code using a more convenient graphical tool - an Integrated Development Environment (IDE).
- Programming interfaces define how individual modules within a software application interact among themselves or how the application itself interacts with its users.
- MVC is a software design architecture which divides the application into three interconnected modules: Model (data), View (user interface), and Controller (input/output and data manipulation).
- The software project we use throughout this course is an example of an MVC application that manipulates patients’ inflammation data and performs basic statistical analysis using Python.
