Identifying the anatomy of SummarizedExperiment and QFeatures objects
Overview
Teaching: 30 min
Exercises: 20 minQuestions
How do SummarizedExperiment and QFeatures objects look like?
Objectives
Recognize the anatomy of SummarizedExperiment and QFeatures objects.
Execute common operations on SummarizedExperiment and QFeatures objects.
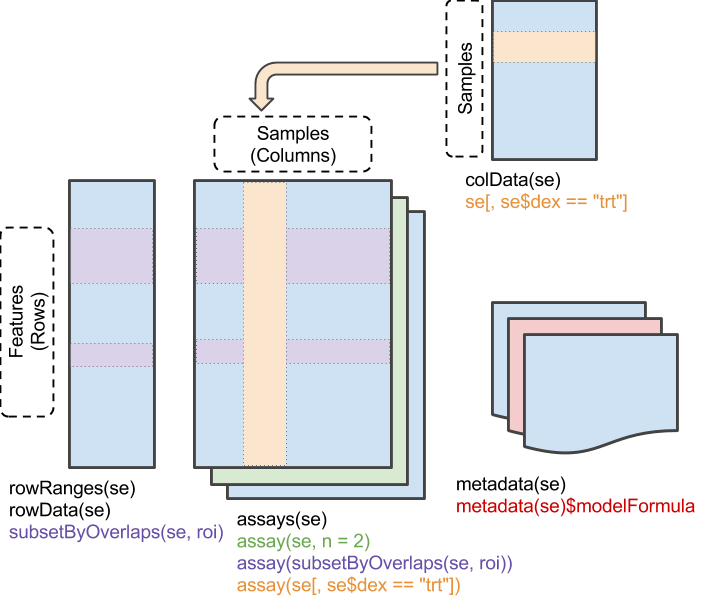
Anatomy of a SummarizedExperiment object
These are the component pieces of the SummarizedExperiment for data representation.
We can access different levels of information of a SummarizedExperiment object using the following functions:
- assay(): A matrix-like or list of matrix-like objects of identical dimension
- colData(): Annotations on each column, as a DataFrame.
- rowData(): Annotations on each row.
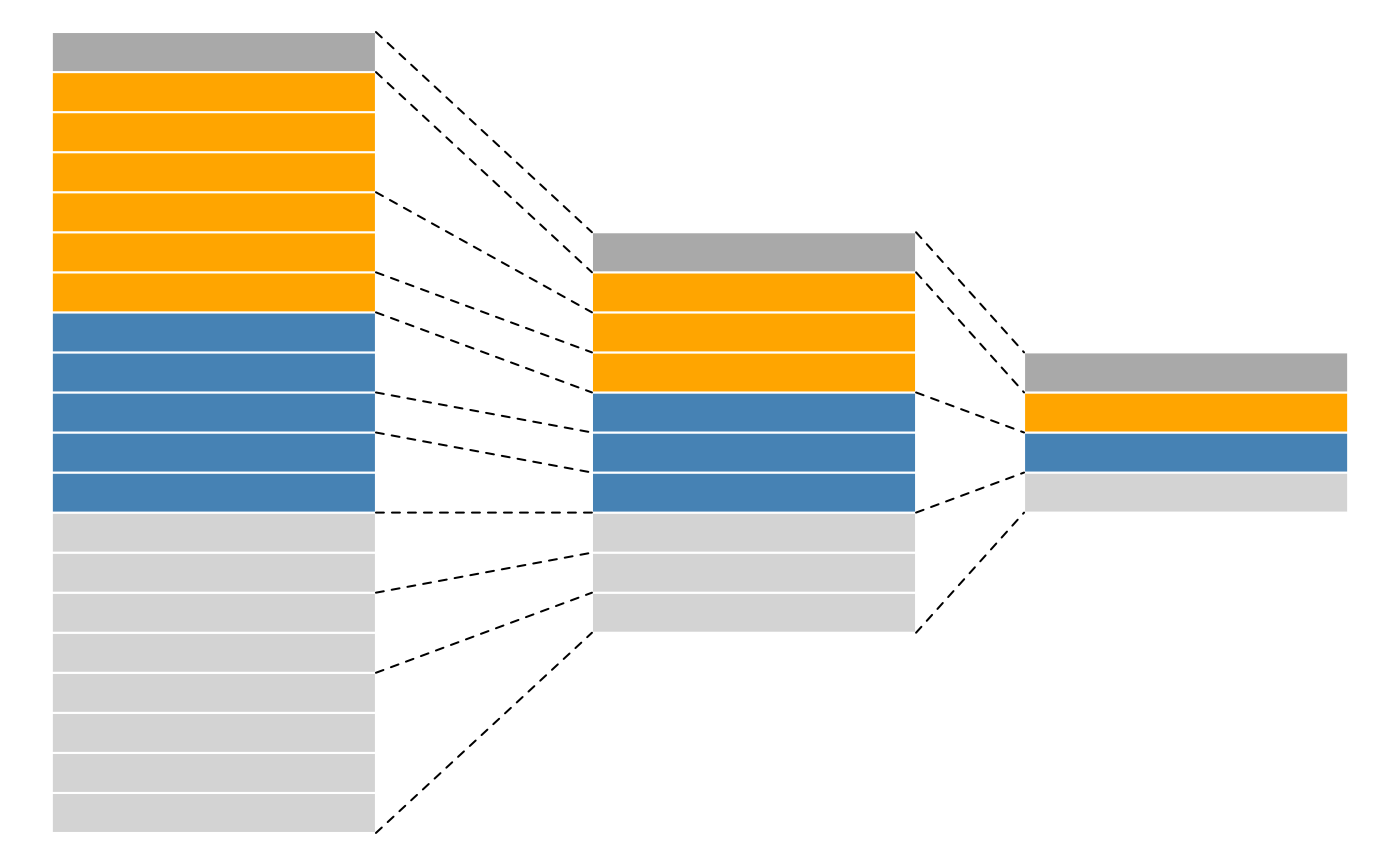
Anatomy of a QFeatures object
QFeatures objects form QFeatures package are based on the SummarizedExperiment and MultiAssayExperiment classes and provides infrastructure to manage and analyse quantitative features from mass spectrometry experiments. It follows a hierarchical structure: spectra (first column in the picture) compose peptides (second column in the picture) which in turn compose proteins (third column in the picture). The main advantage of this structure is that is very easy to navigate across spectra, peptide and protein quantitative data.
Example:
We can load a simplify example feat1 data, which is composed of single assay of class SummarizedExperiment composed of 10 rows and 2
columns.
library("QFeatures")
data(feat1)
feat1
Similarly to what we have done for SummarizedExperiment object , we can extract information from QFeatures
colData(feat1)
assay(feat1[[1]])
rowData(feat1[[1]])
Key Points