Content from Course introduction
Last updated on 2025-07-01 | Edit this page
Overview
Questions
- What is open and reproducible research?
- Why are these practices important, in particular in the context of software used to support such research?
Objectives
- Describe the principles of open and reproducible research and why they are of value in the research community
- Explain how the concept of reproducibility translates into practices for building better research software
- Setup machine with software and data used to teach this course
Software is fundamental to modern research - some of it would even be impossible without software. From short, thrown-together temporary scripts written to help with day-to-day research tasks, through an abundance of complex data analysis spreadsheets, to the hundreds of software engineers and millions of lines of code behind international efforts such as the Large Hadron Collider, there are few areas of research where software does not have a fundamental role.
This course teaches good practices and reproducible working methods that are agnostic of a programming language (although we will use Python code in our examples). It aims to provide researchers with the tools and knowledge to feel confident when writing good quality and sustainable software to support their research. Although the discussion will often focus on software developed in the context of research, most of the good practices introduced here are beneficial to software development more generally.
The lesson is particularly focused on one aspect of good (scientific) software development practice: improving software to enhance reproducibility. That is, enabling others to run our code and obtain the same results we did.
Why should I care about reproducibility?
Scientific transparency and rigor are key factors in research. Scientific methodology and results need to be published openly and replicated and confirmed by several independent parties. However, research papers often lack the full details required for independent reproduction or replication. Many attempts at reproducing or replicating the results of scientific studies have failed in a variety of disciplines ranging from psychology (The Open Science Collaboration (2015)) to cancer sciences (Errington et al (2021)). These are called the reproducibility and replicability crises - ongoing methodological crises in which the results of many scientific studies are difficult or impossible to repeat.
Reproducible research is a practice that ensures that researchers can repeat the same analysis multiple times with the same results. It offers many benefits to those who practice it:
- Reproducible research helps researchers remember how and why they performed specific tasks and analyses; this enables easier explanation of work to collaborators and reviewers.
- Reproducible research enables researchers to quickly modify analyses and figures - this is often required at all stages of research and automating this process saves loads of time.
- Reproducible research enables reusability of previously conducted tasks so that new projects that require the same or similar tasks become much easier and efficient by reusing or reconfiguring previous work.
- Reproducible research supports researchers’ career development by facilitating the reuse and citation of all research outputs - including both code and data.
- Reproducible research is a strong indicator of rigor, trustworthiness, and transparency in scientific research. This can increase the quality and speed of peer review, because reviewers can directly access the analytical process described in a manuscript. It increases the probability that errors are caught early on - by collaborators or during the peer-review process, helping alleviate the reproducibility crisis.
However, reproducible research often requires that researchers implement new practices and learn new tools. This course aims to teach some of these practices and tools pertaining to the use of software to conduct reproducible research.
Review the Reproducible Research Discussion for a more in-depth discussion of this topic.
Practices for building better research software
The practices we will cover for building better research software fall into three areas.
1. Things you can do with your own computing environment to enhance the software
- Using virtual development environments ensures your software can be developed and run consistently across different systems, making it easier for you and others to run, reuse, and extend your code.
2. Things you can do to improve the source code of the software itself
- Organising and structuring your code and project directory keeps your software clean, modular, and reusable, enhancing its readability, extensibility, and reusability.
- Following coding conventions and guides for your programming language ensures that others find it easy to read your code, reuse or extend it in their own examples and applications.
- Testing can save time spent on debugging and ensures that your code is correct and does what it is set out to do, giving you and others confidence in your code and the results it produces.
3. Things you can do to make the software easier for other people to use
- Using version control and collaboration platforms like GitHub, GitLab, and CodeBerg makes it easier to share code and work on it together.
- Fostering a community around your software and promoting collaboration helps to grow a user base for your software and contributes to its long-term sustainability.
- Providing clear and comprehensive documentation, including code comments, API specifications, setup guides, and usage instructions, ensures your software is easy to understand, use, and extend (by you and others).
- Accompanying your software with clear information about its licensing terms and how it should be cited ensures that others can reuse and adapt your code with confidence and that you receive credit when they do so.
Challenge
Individually,
- reflect on what practices (and tools) you are already using in your software development workflow,
- list at least 3 new practices or tools that you would like to start employing or using.
Write your reflections in the shared collaborative document.
Our research software project
You are going to follow a fairly typical experience of a new
researcher (e.g. a PhD student or a postdoc) joining a research group.
You were emailed some spacewalks data and analysis code bundled in the
spacewalks.zip archive, written by another group member who
worked on similar things but has since left. You need to be able to
install and run this code on your machine, check you can understand it
and then adapt it to your own project.
As part of the setup for this
course, you may have downloaded or been emailed the
spacewalks.zip archive. If not, you can download it now.
Save the spacewalks.zip archive to your home directory and
extract it - you should get a directory called
spacewalks.
The first thing you may want to do is inspect the content of the code and data you received. We will use VS Code for browsing, inspecting, modifying files and running our code.
VS Code is a very handy tool for software development and is used by
many researchers worldwide. It “understands” the syntax of many
different file types - for example Python, JSON, CSV, etc. - either
natively or via extensions that can be installed. To open our directory
spacewalks in VS Code – go to File -> Open
Folder and find spacewalks.
You may notice that the software project contains:
-
A JSON file called
data.json- a snippet of which is shown below - with data on extra-vehicular activities (EVAs, i.e. spacewalks) undertaken by astronauts and cosmonauts from 1965 to 2013 (data provided by NASA via its Open Data Portal). JSON data file snippet showing EVA/spacewalk data including EVA id, country, crew members, vehicle type, date of the spacewalk, duration, and purpose)
JSON data file snippet showing EVA/spacewalk data including EVA id, country, crew members, vehicle type, date of the spacewalk, duration, and purpose) -
A Python script called
my code v2.pycontaining some analysis code.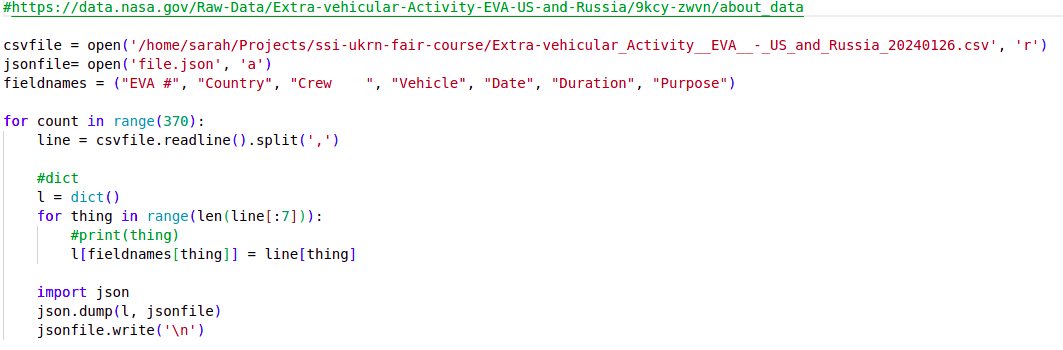 A first few lines of a Python script
A first few lines of a Python scriptThe code in the Python script does some common research tasks:
- Reads in the data from the JSON file
- Changes the data from one data format to another and saves to a file in the new format (CSV)
- Performs some calculations to generate summary statistics about the data
- Makes a plot to visualise the data
Read and understand data and code
Individually inspect the code and try and see if you can understand what the code is doing and how it is organised.
In the shared document, write down anything that you think is not “quite right”, not clear, is missing, or could be done better.
Below are some suggested questions to help you assess the code. These are not the only criteria on which you could evaluate the code and you may find other aspects to comment on.
- If these files were emailed to you, or sent on a chat platform, or handed to you on a memory stick, how easy would it be to find them again in 6 months, or 3 years?
- Can you understand the code? Does it make sense to you?
- Could you run the code on your platform/operating system (is there documentation that covers installation instructions)? What programs or libraries do you need to install to make it work (and which versions)? Are these commonly used tools in your field?
- Are you allowed to use the code in your own research? If you did, would your collaborators expect credit in some form (paper authorship, citation or acknowledgement)? Are you allowed to edit the files? Publish them? Share them with others?
- Is the code written in a way that allows you to easily modify or extend it? How easy would it be to change its parameters to calculate a different statistic, or run on a different input file?
This is a (non-exhaustive) list of things that could be fixed/improved with our code and data:
File and variable naming
- data (
data.json) and Python script (my code v2.py) files could have more descriptive names - Python script (
my code v2.py) should not contain blank spaces as it may cause problems when running the code from command line - variables (e.g.
w) should have more descriptive and meaningful names - version control is embedded in file name - there are better ways of keeping track of changes to code and its different versions
Code organisation and style
- import statements should be grouped at the top
- commenting and uncommenting code should not be used to direct the flow of execution / type of analysis being done
- the code lacks comments, documentation and explanations
- code structure could be improved to be more modular and not one monolithic piece of code - e.g. use functions for reusable units of functionality
- unused variables (e.g.
fieldnamesmeant to be used when saving data to CSV file) are polluting the code and confusing the person reading the code - spaces should not be used in column names as it can lead to error when reading the data in
Code content and correctness
- fixing the loop to 374 data entries is not reusable on other data files and would likely break if the data file changed
- running the code twice causes the program to fail as the result file from the previous run will exist (which the code does not check for) and the script will refuse to overwrite it
- the code does not specify the encoding when reading the data in, and we are also not sure what encoding the data was saved in originally
- how can we be confident the data analysis and plot that is produced as a result are correct?
Documentation
- there is no README documentation to orient the user
- there is no licence information to say how the code can be reused (which then means it cannot be reused at all)
- it is not clear what software dependencies the code has
- there are no installation instructions or instructions on how to run the code
As you have seen from the previous exercise - there are quite a few things that can be improved with this code. We will try to make this research software project a “bit better” for future use.
Let’s check your setup now to make sure you are ready for the rest of this course.
Check your setup
From a command line terminal on your operating system or within VS Code run the following commands to check you have installed all the tools listed in the Setup page and that are functioning correctly.
Checking the command line terminal:
$ date$ echo $SHELL$ pwd$ whoami
Checking Python:
$ python --version$ python3 --version$ which python$ which python3
Checking Git and GitHub:
$ git --help$ git config --list$ ssh -T git@github.com
Checking VS Code:
$ code$ code --list-extensions
The expected out put of each command is:
- Today’s date
-
bashorzsh- this tells you what shell language you are using. In this course we show examples in Bash. - Your “present working directory” or the folder where your shell is running
- Your username
- In this course we are using Python 3. If
python --versiongives you Python 2.x you may have two versions of Python installed on your computer and need to be careful which one you are using. - Use this command to be certain you are using Python version 3, not 2, if you have both installed.
- The file path to where the Python version you are calling is installed.
- If you have more than one version these should be different paths, if both 5. and 6. gave the same result then 7. and 8. should match as well.
- The help message explaining how to use the
gitcommand. - You should have
user.name,user.emailandcore.editorset in your Git configuration. Check that the editor listed is one you know how to use. - This checks if you have set up your connection to GitHub correctly.
If is says
permission deniedyou may need to look at the instructions for setting up SSH keys again on the Setup page. - This should open VSCode in your current working directory. macOS users may need to first open VS Code and add it to the PATH.
- You should have the extensions GitLens, Git Graph, Python, JSON and Excel Viewer installed to use in this course.
Further reading
We recommend the following resources for some additional reading on reproducible research:
- Reproducible research through reusable code is a one day course by the Netherlands eScience Center which discusses similar themes to this course, but in a shorter format.
- The Turing Way’s “Guide for Reproducible Research”
- A Beginner’s Guide to Conducting Reproducible Research, Jesse M. Alston, Jessica A. Rick, Bulletin of The Ecological Society of America 102 (2) (2021), https://doi.org/10.1002/bes2.1801
- “Ten reproducible research things” tutorial
- FORCE11’s FAIR 4 Research Software (FAIR4RS) Working Group
- “Good Enough Practices in Scientific Computing” course
- Reproducibility for Everyone’s (R4E) resources, community-led education initiative to increase adoption of open research practices at scale
- Training materials on different aspects of research software engineering (including open source, reproducibility, research software testing, engineering, design, continuous integration, collaboration, version control, packaging, etc.), compiled by the INTERSECT project
- Curated resources by the Framework for Open and Reproducible Research Training (FORRT)
Acknowledgements and references
The content of this course borrows from or references various work.
Content from Better start with a software project
Last updated on 2025-07-01 | Edit this page
Overview
Questions
- What is a version control system?
- Why is version control essential to building good software
- What does a standard version control workflow look like?
Objectives
- Set up version control for our software project to track changes to it
- Create self-contained commits using Git to incrementally save work
- Push new work from a local machine to a remote server on GitHub
In this episode, we will set up our new research software project using some good practices from the start. This will lay the foundation for long-term sustainability of our code, collaboration, and reproducibility.
This starts with following naming conventions for files, employing version control, and (in the next episode) setting up a virtual development environment with software dependencies to ensure the project can be more easily and reliably run, shared and maintained. Next (over the rest of the course) - adding tests, setting up automation (e.g. continuous integration), documenting software and including metadata such as licensing, authorship and citation will ensure the results our software produces can be trusted and others can build upon it with confidence.
Let’s begin by creating a new software project from our existing code, and start tracking changes to it with version control.
From script to software project
In the previous episode you have unzipped spacewalks.zip
into a directory spacewalks in your home directory. If you
have not opened the software directory in VS Code already – go to
File -> Open Folder and find
spacewalks.
We also need access to a command line terminal to type various commands. In VS Code start a new terminal via Terminal -> New Terminal (Windows users need to make sure the new terminal is “GitBash”; not “PowerShell” or “cmd”). Alternatively, you can work with a shell terminal directly (and not within VS Code), if you so prefer.
If you are not already inside this directory, from your command line terminal you can navigate to it and list its contents with:
BASH
cd ~/spacewalks
ls -la
total 272
drwx------@ 5 mbassan2 staff 160 26 Jun 11:35 .
drwx------@ 489 mbassan2 staff 15648 26 Jun 11:41 ..
drwxrwxr-x@ 4 mbassan2 staff 128 4 Apr 10:48 astronaut-data-analysis-old
-rw-rw-r--@ 1 mbassan2 staff 132981 4 Apr 10:48 data.json
-rw-rw-r--@ 1 mbassan2 staff 1518 4 Apr 10:48 my code v2.pyOver the rest of the course, we will transform a collection of these files into a well-structured software project that follows established good practices in research software engineering.
The first thing you may notice that our software project contains
folder astronout-data-analysis-old which presumably tries
to keep track of older versions of the code. There is a better way to do
that using version control tool, such as Git, and we can delete this
folder so it does not cause confusion:
Version control
Before we do any further changes to our software, we want to make sure we can keep a history of what changes we have done since we inherited the code from our colleague.
We can track changes with version control. Later on, we will store those changes on a remote server too – both for safe-keeping and to make them easier to share with others. In later episodes, we will also see how version control makes it easier for multiple collaborators to work together on the same project at the same time and combine their contributions.
Version control refresher
What is a version control system?
Version control systems are tools that let you track changes in files over time in a special database that allows users to “travel through time”, and compare earlier versions of the files with the current state. Think of a version control system like turning on ‘Track Changes’ on Microsoft Word/Google Docs, but for any files you want, and a lot more powerful and flexible.
Why use a version control system?
As scientists, our main motivation for using version control is reproducibility. By tracking and storing every change we make, we can restore our project to the state it was at any point in time. This is incredibly useful for reproducing results from a specific version of the code, or tracking down which changes we (or others) made introduced bugs or changed our results.
The other benefit of version control is it provides us with a history of our development. As we make each change, we record what it was, and why we made it. This helps make our development process transparent and auditable – which is a good scientific practice.
It also makes our project more sustainable - as our data, software and methods (knowledge) remain usable and accessible over time (especially if made available in shared version controlled code repositories), even after the original funding ends or team members move on.
Git version control system
Git is the most popular version control system used
by researchers worldwide, and the one we’ll be using. Git is used mostly
for managing code when developing software, but it can track
any files – and is particularly effective with text-based files
(e.g. source code like .py, .c,
.r, but also .csv, .tex and
more).
Git helps multiple people work on the same project (even the same file) at the same time. Initially, we will use Git to start tracking changes to files on our local machines; later on we will start sharing our work on GitHub allowing other people to see and contribute to our work.
Git refresher
Git stores files in repositories - directories where changes to the files can be tracked. The diagram below shows the different parts of a Git repository, and the most common commands used to work with one.
-
Working directory - a local directory (including
any subdirectories) where your project files live, and where you are
currently working. It is also known as the “untracked” area of Git. Any
changes to files will be marked by Git in the working directory. Git
will only save changes that you explicitly tell it to. Using
git add FILENAMEcommand, can you tell Git to start tracking changes to fileFILENAMEin your working directory. -
Staging area (index) - once you tell Git to start
tracking changes to files (with
git add FILENAMEcommand), Git saves those changes in the staging area on your local machine. Each subsequent change to the same file needs to be followed by anothergit add FILENAMEcommand to tell Git to update it in the staging area. To see what is in your working directory and staging area at any moment (i.e. what changes is Git tracking), you can run the commandgit status. The staging area lets you bundle together groups of changes to save to your repository. -
Local repository - stored within the
.gitdirectory of your project locally, this is where Git wraps together all your changes from the staging area and puts them using thegit commitcommand. Each commit is a new, permanent snapshot (checkpoint, record) of your project in time, which you can share or revert to. -
Remote repository - this is a version of your
project that is hosted somewhere on the Internet (e.g., on GitHub,
GitLab or somewhere else). While your project is nicely
version-controlled in your local repository, and you have snapshots of
its versions from the past, if your machine crashes you still may lose
all your work. Plus, sharing or collaborating on local work with others
requires lots of emailing back and forth. Working with a remote
repository involves ‘pushing’ your local changes to it (using
git push), and pulling other people’s changes back to your local copy (usinggit fetchorgit pull). This keeps the two in sync in order to collaborate, with a bonus that your work also gets backed up to another machine. Best practice when collaborating with others on a shared repository is to always do agit pullbefore agit push, to ensure you have any latest changes before you push your own.
Start tracking changes with Git
Before we start, if you forgot to do it during setup, tell Git to use
main as the default branch. More modern versions of Git use
main, but older ones still use master as their
main branch. They work the same, but we want to keep things consistent
for clarity.
At this point, we should be located in our spacewalks
directory. We want to tell Git to make spacewalks a
repository – a directory where Git can track changes to our files. We do
that with:
We can check everything is set up correctly by asking Git to tell us the status of our project:
OUTPUT
On branch main
No commits yet
Untracked files:
(use "git add <file>..." to include in what will be committed)
data.json
my code v2.py
nothing added to commit but untracked files present (use "git add" to track)This tells us that Git has noticed two files in our directory, but unlike Dropbox or OneDrive, it does not automatically track them. We need to tell Git explicitly which files we want it to track. This is not a handicap, but rather helpful, since scientific code can have vast inputs or outputs we might not want Git to track and store (GBs ot TBs of space telescope data) or require sensitive information we cannot share (for example, medical records).
Add files into repository
We can tell Git to track a file using git add:
and then check the right thing happened:
OUTPUT
On branch main
No commits yet
Changes to be committed:
(use "git rm --cached <file>..." to unstage)
new file: data.json
new file: my code v2.pyGit now knows that should track the changes to
my code v2.py and data.json, but it has not
‘committed’ those changes to the record yet. A commit is a snapshot of
how your tracked files have changed at a stage in time. To create a
commit that records we added two new files, we need to run one more
command:
OUTPUT
[main (root-commit) bf55eb7] Add the initial spacewalks data and code
2 files changed, 437 insertions(+)
create mode 100644 data.json
create mode 100644 my code v2.pyAt this point, Git has taken everything we have told it to save with
the git add command and stored a copy (snapshot) of the
files in a special, hidden .git directory. This is called a
commit (or revision).
The -m option means message, and records a short,
descriptive, and specific comment that will help us remember later on
what we did and why. If we run git commit without
-m , Git will still expect a message – and will launch a
text editor so that we can write a longer one.
Remember, good commit messages start with a brief (<50 characters) statement about the changes made in the commit. Generally, the message should complete the sentence “If applied, this commit will…”. If you want to go into more detail, add a blank line between the summary line and your additional notes. Use this additional space to explain why you made changes and/or what their impact will be.
If we run git status now, we see:
OUTPUT
On branch main
nothing to commit, working tree cleanThis tells us that everything is up to date.
Where are my changes?
If we run ls at this point, we’ll still only see two
files – our script, and our dataset. Git saves information about our
files’ history in the special .git directory mentioned
earlier. This both stops our folders being cluttered with old versions,
and also stops us accidentally deleting them!
You can see the hidden Git directory using the -a flag
to show all files and folders:
OUTPUT
.
..
.gitIf you delete it, your directory will stop being a repository, and it
will lose your history of changes. You never need to look into
.git yourself – Git adds useful commands to do that, which
are covered later on.
Make a change
You may have noticed that the script we received contain blank spaces
in filename. This meant that, when we were typing the script’s name into
the terminal, we had to add a slash before the space like this:
my\ code\ v2.py. Using a backslash in this way is called
“escaping”. It lets the terminal know to treat the space as part of the
filename, and not a separate argument. It is a bit inconvenient and can
cause problems if you forget, so best practise is to avoid spaces in
filenames. The simplest fix is to replace the spaces with underscores
_ instead.
To rename the files using git you can use the git mv
command:
If you run git status again, you’ll see Git has noticed
the change in the filename. Note, git mv handles the name
change directly, instead of seeing a deleted file and a new file as
would be the case if we’d used mv and then
git add. It also stages the changes to be committed.
OUTPUT
On branch main
Changes to be committed:
(use "git restore --staged <file>..." to unstage)
renamed: my code v2.py -> my_code_v2.pyRename our data and output files
Now that we know how to rename files in Git, we can use it to make our files and code a bit easier to understand.
We may want to:
- Give our script and input data file more meaningful names, e.g
eva_data_analysis.pyandeva-data.json. This change also uses removes version tracking from the script name as we are using Git for version control any more as Git will keep track of that for us. - Choose informative file names for our output data file
(e.g.
eva-data.csv) and plot (cumulative_eva_graph.png). - Use relative paths (e.g.
./eva-data.json) instead of absolute paths (e.g.home/sarah/Projects/ssi-ukrn-fair-course/data.csv) to the files (which were hardcoded to the path on our colleagues machine and would not work on ours). - Update the Python script with these changes.
Update the filenames in the repo
Try to make these changes yourself.
- Give our Python script and input data file informative names -
eva_data_analysis.pyandeva-data.json, respectively. - Update other file names and paths used in the script - output CSV
data (
eva-data.csvto match the new input data name) and plot(cumulative_eva_graph.png). - Stage and commit these changes in the Git repository.
Firstly, let’s update the file names in our Python script from VS Code:
PYTHON
data_f = open('./eva-data.json', 'r')
data_t = open('./eva-data.csv','w')
g_file = './cumulative_eva_graph.png'Next, we need to rename the files on the file system using Git:
BASH
git mv data.json eva-data.json
git mv my_code_v2.py eva_data_analysis.py
git add eva_data_analysis.py
git statusOUTPUT
On branch main
Changes to be committed:
(use "git restore --staged <file>..." to unstage)
renamed: data.json -> eva-data.json
renamed: my_code_v2.py -> eva_data_analysis.pyFinally, we can commit our changes:
Interacting with a remote Git server
Git is distributed version control system and lets us synchronise work between multiple copies of the same repository - which may not be on your machine (called remote repositories). So far, we have used a local repository on our machine and, even though we have been incrementally saving our work in a way that is recoverable, if we lost our machine then we would lose all our code along with it,
Fortunately, we can easily upload our local repository, with all our code and the history of our development, to a remote server so that it can be backed-up and recovered in future.
GitHub is an online software development platform that can act as a central remote server. It uses Git, and provides facilities for storing, tracking, and collaborating on software projects. Other Git hosting services are available, such as GitLab and Bitbucket.
Putting our projects on GitHub helps protect them against deletion, and also makes it easy to collaborate and share them. Our collaborators can access the project, download their own copy, and even contribute work back to us.
Let’s push our local repository to GitHub and share it publicly.
In your browser, navigate to https://github.com and sign into your account.
-
In the top right hand corner of the screen, there is a menu labelled “+” with a dropdown. Click the dropdown and select “New repository” from the options:
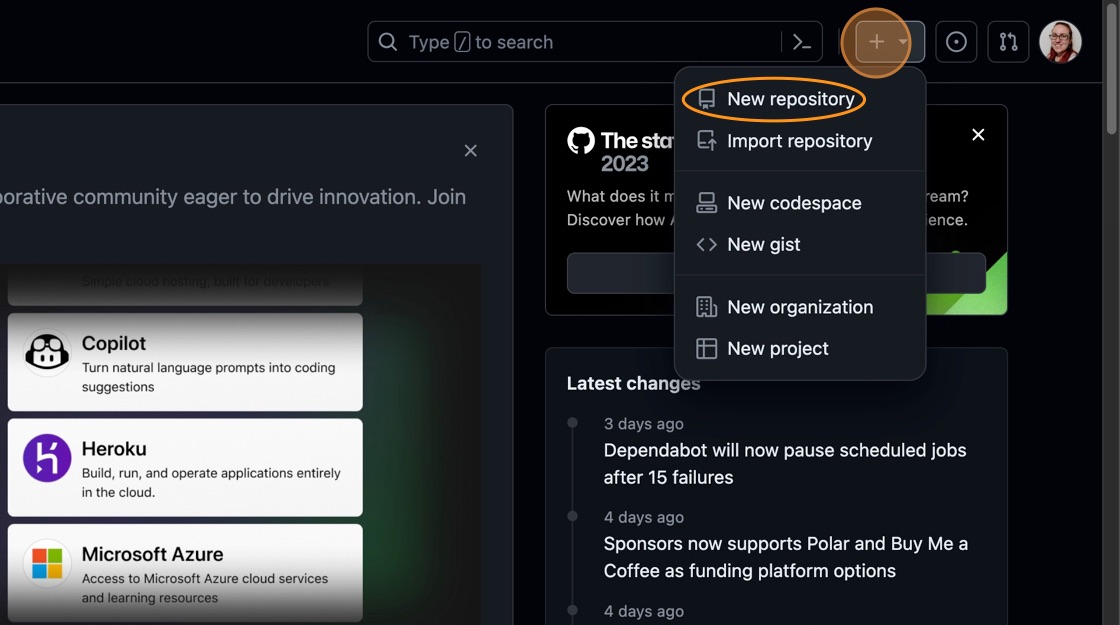 Creating a new GitHub repository
Creating a new GitHub repository -
You will be presented with some options to fill in or select while creating your repository. In the “Repository Name” field, type “spacewalks”. This is the name of your project and matches the name of your local folder.
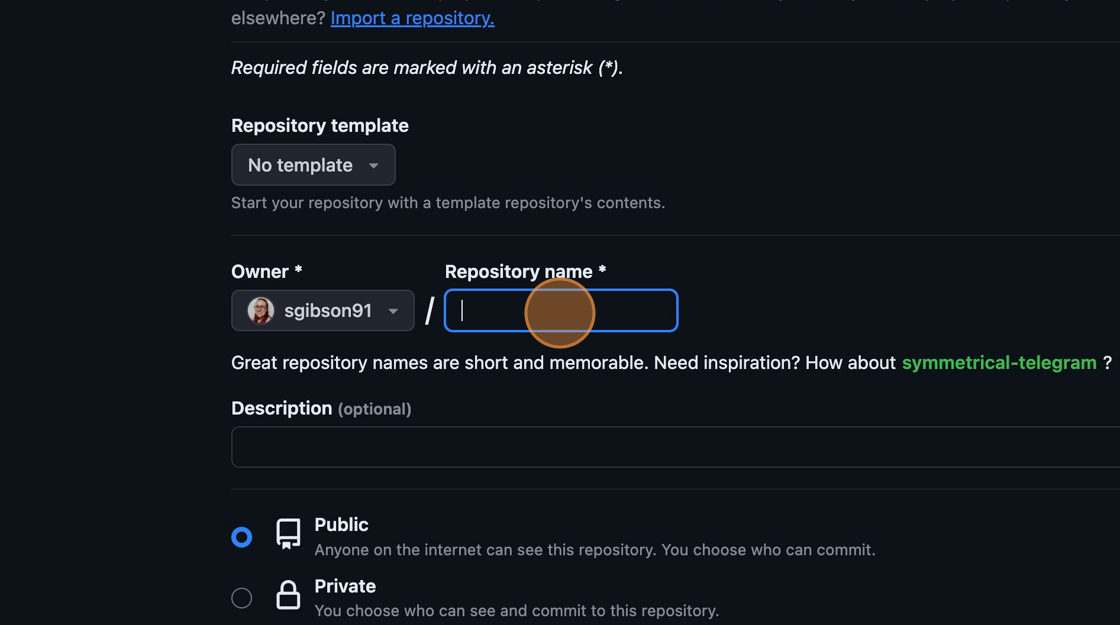 Naming the GitHub repository
Naming the GitHub repositoryEnsure the visibility of the repository is “Public” and leave all other options blank. Since this repository will be connected to a local repository, it needs to be empty which is why we chose not to initialise with a README or add a license or
.gitignorefile. Click “Create repository” at the bottom of the page: Complete GitHub repository creation
Complete GitHub repository creation -
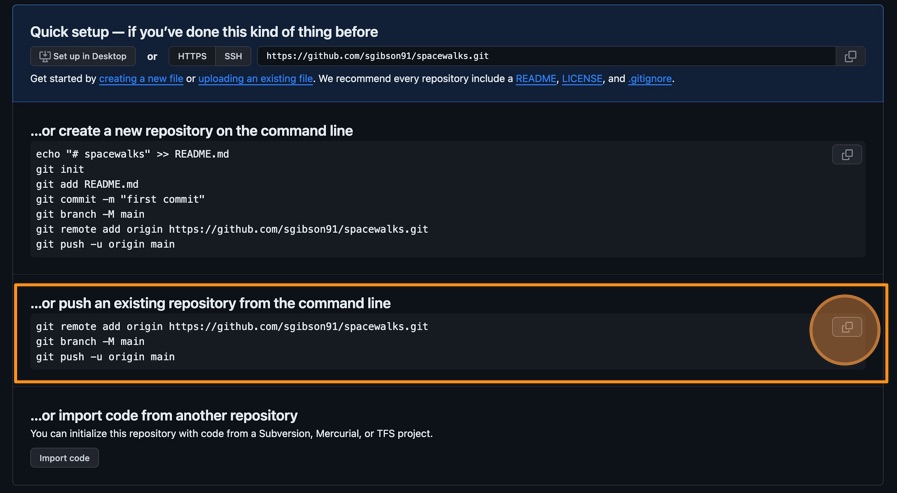
Now we have a remote repository on GitHub’s servers, you need to send it the files and history from your local repository. GitHub provides some instructions on how to do that for different scenarios. Change the toggle on the right side from “HTTPS” to “SSH”, then look at the heading “…or push an existing repository from the command line”. You should see instructions that look like this:
BASH
git remote add origin git@github.com/<YOUR_GITHUB_HANDLE>/spacewalks.git git branch -M main git push -u origin mainIt is very important you make sure you switch from “HTTPS” to “SSH”. In the setup, we configured our GitHub account and our local machine for SSH. If you select HTTPS, you will not be able to upload your files.
You can copy these commands using the button that looks like two overlapping squares to the right-hand side of the commands. Paste them into your terminal and run them.
- If you refresh your browser window, you should now see the two files
eva_data_analysis.pyandeva-data.jsonvisible in the GitHub repository, matching what you have locally on your machine.
If you were wondering about what those commands did, here is the explanation.
This command tells Git to create a remote called
“origin” and link it to the URL of your GitHub repository. A
remote is a version control concept where two (or more)
repositories are connected to each other, in such a way that they can be
kept in sync by exchanging commits. “origin” is a name used to refer to
the remote repository. It could be called anything, but “origin” is a
common convention for Git since it shows which is considered the “source
of truth”. This is particularly useful when many people are
collaborating on the same repository.
git branch is a command used to manage branches. We’ll
discuss branches later on in the course. We saw this command during
setup and earlier in this episode - it ensures the branch we are working
on is called “main”. This will be the default branch of the project for
everyone working on it.
The git push command is used to update a remote
repository with changes from your local repository. This command tells
Git to update the “main” branch on the “origin” remote. The
-u flag (short for --set-upstream) sets the
‘tracking reference’ for the current branch, so that in future
git push will default to sending to
origin main.
Summary
We have created a new software project and used version control system Git to track changes to it. We can now look back at our work, compare different code versions, and even recover past states. We have also published our software to a remote repository located on GitHub, where it is both secure and shareable.
These skills are critical to reproducible and sustainable science. Software is science, and being able to share the specific version of code used in a paper is required for reproducibility. But we, as researchers, also benefit from having a clear, self-documented record of what we did, when and why. It makes it much easier to track down and fix our bugs, return to work on projects we took a break from, and even for other people to pick up our work.
Before we start making changes to the code, we have to set up a development environment with software dependencies for our project to ensure this metadata about our project is recorded and shared with anyone wishing to download, run or extend our software (and this includes ourselves on a different machine or operating system).
Further reading
We recommend the following resources for some additional reading on the topic of this episode:
- Software Carpentry’s Git Novice lesson
- The Turing Way’s “Guide to Version Control”
- “How Git Works” course on Pluralsight
- How to Write a Good Commit Message
- Git Commit Good Practice
Also check the full reference set for the course.
Key Points
- Version control systems are software that tracks and manages changes to a project over time
- Using version control aids reproducibility since the exact state of the software that produced an output can be recovered
- A commit represents the smallest unit of change to a project
- Commit messages describe what each commit contains and should be descriptive
- GitHub is a hosting service for sharing and collaborating on software
- Using version control is one of the first steps to creating a software project from a bunch of scripts - by investing in these practices early, researchers can create software that supports their work more effectively and enables others to build upon it with confidence.
Content from Reproducible software environments
Last updated on 2025-07-01 | Edit this page
Overview
Questions
- What are virtual environments in software development and why use them?
- How can we manage Python virtual coding environments and external (third-party) libraries on our machines?
Objectives
- Set up a Python virtual coding environment for a software project
using
venvandpip.
So far we have created a local Git repository to track changes in our software project and pushed it to GitHub to enable others to see and contribute to it. We now want to start developing the code further.
Software dependencies
If we have a look at our script, we may notice a few
import lines such as: import json,
import csv, import datetime as dt and
import matplotlib.pyplot as plt throughout the code. This
means that our code depends on or requires several external
libraries (also called third-party packages or
dependencies) to function - namely json,
csv, datetime and matplotlib.
Python applications often use external libraries that do not come as part of the standard Python distribution. This means that you will have to use a package manager tool to install them on your system. Applications will also sometimes need a specific version of an external library (e.g. because they were written to work with feature, class, or function that may have been updated in more recent versions), or a specific version of Python interpreter. This means that each Python application you work with may require a different setup and a set of dependencies so it is useful to be able to keep these configurations separate to avoid confusion between projects. The solution for this problem is to create a self-contained virtual environment per project, which contains a particular version of Python installation plus a number of additional external libraries.
What are virtual software environments?
So what exactly are virtual software environments, and why use them?
A Python virtual environment helps us create an isolated working copy of a software project that uses a specific version of Python interpreter together with specific versions of a number of external libraries installed into that virtual environment. Python virtual environments are implemented as directories with a particular structure within software projects, containing links to specified dependencies allowing isolation from other software projects on your machine that may require different versions of Python or external libraries.
It is recommended to create a separate virtual environment for each project. Then you do not have to worry about changes to the environment of the current project you are working on affecting other projects - you can use different Python versions and different versions of the same third party dependency by different projects on your machine independently from one another.
We can visualise the use of virtual environments for different Python
projects on the same machine as follows: 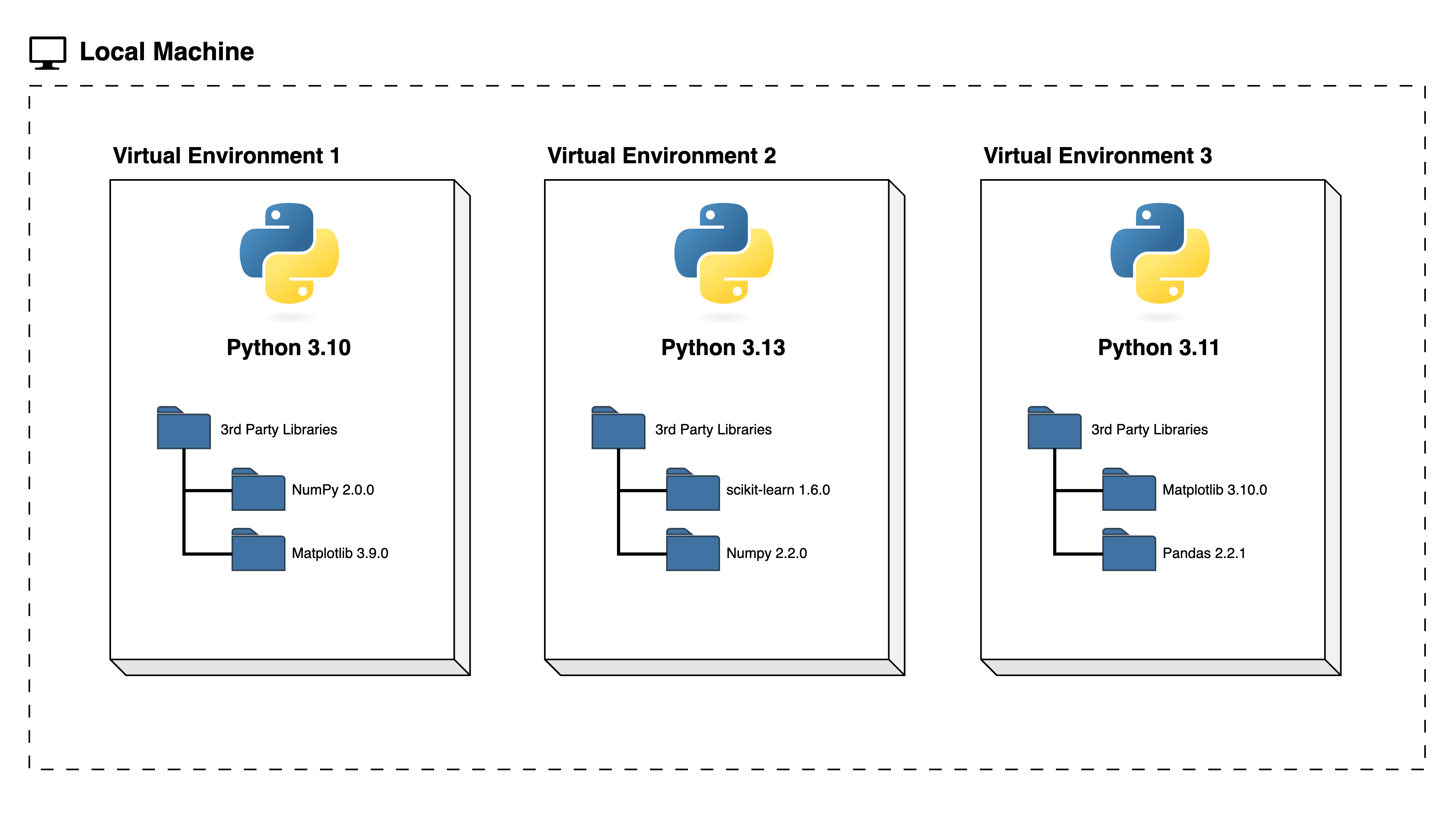
Another big motivator for using virtual environments is that they make sharing your code with others much easier - as we will see shortly you can record your virtual environment in a special file and share it with your collaborators who can then recreate the same development environment on their machines.
You do not have to worry too much about specific versions of external libraries that your project depends on most of the time. Virtual environments also enable you to always use the latest available version without specifying it explicitly. They also enable you to use a specific older version of a package for your project, should you need to.
Truly reproducible environments are difficult to attain
Creating and managing isolated environments for each of your software projects and sharing descriptions of those environments alongside the relevant code is a great way to make your software and analyses much more reproducible. However, “true” computational reproducibility is very difficult to achieve. For example, the tools we will use in this lesson only track the dependencies of our software, remaining unanware of other aspects of the software’s environment such as the operating system and hardware of the system it is running on. These properties of the environment can influence the running of the software and the results it produces and should be accounted for if a workflow is to be truly reproducible.
Although there is more that we can do to maximise the reproducibility of our software/workflows, the steps described in this episode are an excellent place to start. We should not let the difficulty of attaining “perfect” reproducibility prevent us from implementing “good enough” practices that make our lives easier and are much better than doing nothing.
Managing virtual environments
There are several command line tools used for managing Python virtual
environments - we will use venv, available by default from
the standard Python distribution since
Python 3.3.
Part of managing your (virtual) working environment involves
installing, updating and removing external packages on your system. The
Python package manager tool pip is most commonly used for
this - it interacts and obtains the packages from the central repository
called Python Package Index (PyPI).
So, we will use venv and pip in combination
to help us create and share our virtual development environments.
Creating virtual environments
Creating a virtual environment with venv is done by
executing the following command:
where /path/to/new/virtual/environment is a path to a
directory where you want to place it - conventionally within your
software project so they are co-located. This will create the target
directory for the virtual environment.
For our project let’s create a virtual environment called “venv_spacewalks” from our project’s root directory.
Firstly, ensure you are located within the project’s root directory:
If you list the contents of the newly created directory “venv_spacewalks”, on a Mac or Linux system (slightly different on Windows as explained below) you should see something like:
OUTPUT
total 8
drwxr-xr-x 12 alex staff 384 5 Oct 11:47 bin
drwxr-xr-x 2 alex staff 64 5 Oct 11:47 include
drwxr-xr-x 3 alex staff 96 5 Oct 11:47 lib
-rw-r--r-- 1 alex staff 90 5 Oct 11:47 pyvenv.cfgSo, running the python3 -m venv venv_spacewalks command
created the target directory called “venv_spacewalks” containing:
-
pyvenv.cfgconfiguration file with a home key pointing to the Python installation from which the command was run, -
binsubdirectory (calledScriptson Windows) containing a symlink of the Python interpreter binary used to create the environment and the standard Python library, -
lib/pythonX.Y/site-packagessubdirectory (calledLib\site-packageson Windows) to contain its own independent set of installed Python packages isolated from other projects, and - various other configuration and supporting files and subdirectories.
Once you’ve created a virtual environment, you will need to activate it.
On Mac or Linux, it is done as:
On Windows, recall that we have Scripts directory
instead of bin and activating a virtual environment is done
as:
Activating the virtual environment will change your command line’s prompt to show what virtual environment you are currently using (indicated by its name in round brackets at the start of the prompt), and modify the environment so that running Python will get you the particular version of Python configured in your virtual environment.
You can verify you are using your virtual environment’s version of
Python by checking the path using the command which:
When you’re done working on your project, you can exit the environment with:
If you’ve just done the deactivate, ensure you
reactivate the environment ready for the next part:
Note that, since our software project is being tracked by Git, the newly created virtual environment will show up in version control - we will see how to handle it using Git in one of the subsequent episodes.
Installing external packages
We noticed earlier that our code depends on four external
packages/libraries - json, csv,
datetime and matplotlib. As of Python 3.5,
Python comes with in-built JSON and CSV libraries - this means there is
no need to install these additional packages (if you are using a fairly
recent version of Python), but you still need to import them in any
script that uses them. However, we still need to install packages
datetime and matplotlib as they do not come as
standard with Python distribution.
To install the latest version of packages datetime and
matplotlib with pip you use pip’s
install command and specify the package’s name, e.g.:
BASH
(venv_spacewalks) $ python3 -m pip install datetime
(venv_spacewalks) $ python3 -m pip install matplotlibor like this to install multiple packages at once for short:
The above commands have installed packages datetime and
matplotlib in our currently active
venv_spacewalks environment and will not affect any other
Python projects we may have on our machines.
If you run the python3 -m pip install command on a
package that is already installed, pip will notice this and
do nothing.
To install a specific version of a Python package give the package
name followed by == and the version number,
e.g. python3 -m pip install matplotlib==3.5.3.
To specify a minimum version of a Python package, you can do
python3 -m pip install matplotlib>=3.5.1.
To upgrade a package to the latest version,
e.g. python3 -m pip install --upgrade matplotlib.
To display information about a particular installed package do:
OUTPUT
Name: matplotlib
Version: 3.9.0
Summary: Python plotting package
Home-page:
Author: John D. Hunter, Michael Droettboom
Author-email: Unknown <matplotlib-users@python.org>
License: License agreement for matplotlib versions 1.3.0 and later
=========================================================
...
Location: /opt/homebrew/lib/python3.11/site-packages
Requires: contourpy, cycler, fonttools, kiwisolver, numpy, packaging, pillow, pyparsing, python-dateutil
Required-by:To list all packages installed with pip (in your current
virtual environment):
OUTPUT
Package Version
--------------- -----------
contourpy 1.2.1
cycler 0.12.1
DateTime 5.5
fonttools 4.53.1
kiwisolver 1.4.5
matplotlib 3.9.2
numpy 2.0.1
packaging 24.1
pillow 10.4.0
pip 23.3.1
pyparsing 3.1.2
python-dateutil 2.9.0.post0
pytz 2024.1
setuptools 69.0.2
six 1.16.0
zope.interface 7.0.1To uninstall a package installed in the virtual environment do:
python3 -m pip uninstall <package-name>. You can also
supply a list of packages to uninstall at the same time.
Sharing virtual environments
You are collaborating on a project with a team so, naturally, you
will want to share your environment with your collaborators so they can
easily ‘clone’ your software project with all of its dependencies and
everyone can replicate equivalent virtual environments on their
machines. pip has a handy way of exporting, saving and
sharing virtual environments.
To export your active environment - use
python3 -m pip freeze command to produce a list of packages
installed in the virtual environment. A common convention is to put this
list in a requirements.txt file in your project’s root
directory:
BASH
(venv_spacewalks) $ python3 -m pip freeze > requirements.txt
(venv_spacewalks) $ cat requirements.txtOUTPUT
contourpy==1.2.1
cycler==0.12.1
DateTime==5.5
fonttools==4.53.1
kiwisolver==1.4.5
matplotlib==3.9.2
numpy==2.0.1
packaging==24.1
pillow==10.4.0
pyparsing==3.1.2
python-dateutil==2.9.0.post0
pytz==2024.1
six==1.16.0
zope.interface==7.0.1The first of the above commands will create a
requirements.txt file in your current directory. Yours may
look a little different, depending on the version of the packages you
have installed, as well as any differences in the packages that they
themselves use.
The requirements.txt file can then be committed to a
version control system (we will see how to do this using Git in a
moment) and get shipped as part of your software and shared with
collaborators and/or users.
Note that you only need to share the small
requirements.txt file with your collaborators - and not the
entire venv_spacewalks directory with packages contained in
your virtual environment. We need to tell Git to ignore that directory,
so it is not tracked and shared - we do this by creating a file
.gitignore in the root directory of our project and adding
a line venv_spacewalks to it.
BASH
(venv_spacewalks) $ echo "venv_spacewalks/" >> .gitignore
(venv_spacewalks) $ git add .gitignore
(venv_spacewalks) $ git commit -m "ignoring venv folder"Let’s now put requirements.txt under version control and
share it along with our code.
BASH
(venv_spacewalks) $ git add requirements.txt
(venv_spacewalks) $ git commit -m "Initial commit of requirements.txt."
(venv_spacewalks) $ git push origin mainYour collaborators or users of your software can now download your
software’s source code and replicate the same virtual software
environment for running your code on their machines using
requirements.txt to install all the necessary depending
packages.
To recreate a virtual environment from requirements.txt,
from the project root one can do the following:
Another challenge in (long-term) reproducibility
For people (including your future self) to be able to reproduce software environments described in this way, the listed dependencies need to remain available to download and possible to install on the user’s system. These are reasonably safe assumptions for widely-used, actively maintained tools on commonly-used operating systems in the short- to medium-term. However, it becomes less likely that we will be able to recreate such environments as system architectures evolve over time and maintainers stop supporting older versions of software. To achieve this kind of long-term reproducibility, you will need to explore options to provide the actual environment – with dependencies already included – alongside your software e.g. via a containerised environment.
As your project grows - you may need to update your environment for a variety of reasons, e.g.:
- one of your project’s dependencies has just released a new version (dependency version number update),
- you need an additional package for data analysis (adding a new dependency), or
- you have found a better package and no longer need the older package (adding a new and removing an old dependency).
What you need to do in this case (apart from installing the new and
removing the packages that are no longer needed from your virtual
environment) is update the contents of the requirements.txt
file accordingly by re-issuing pip freeze command and
propagate the updated requirements.txt file to your
collaborators via your code sharing platform.
Environment management can be troublesome
Software environment management is a difficult thing to get right, which is one reason why the Python community has come up with so many different ways of doing it over the years. (That webcomic is several years old at the time of writing and the Python environment management ecosystem has only become more complicated since.) Unfortunately, even if you try to follow good practices and keep your environments isolated it is possible – perhaps even likely – that you will face difficulties with installing and updating dependencies on your projects in the coming years. Such issues are particularly likely to appear when you upgrade your computer hardware, operating system, and/or interpreter/compiler. As before, this is not a reason to avoid managing your software environments altogether – or to avoid upgrading your hardware, operating system, etc! If you have descriptions of your environments it will always be easier to reproduce them and keep working with your software than if you need to start from scratch. Furthermore, your expertise will develop as you get more practice with managing your software environments, which will equip you well to troubleshoot problems if and when they arise.
Running the code and reproducing results
We are now setup to run our code from the newly created virtual environment:
You should get a pop up window with a graph. However, some (but not all) Windows users will not. You might instead see an error like:
Traceback (most recent call last):
File "C:\Users\Toaster\Desktop\spacewalks\eva_data_analysis.py", line 30, in <module>
w.writerow(data[j].values())
File "C:\Program Files\WindowsApps\PythonSoftwareFoundation.Python.3.12_3.12.2544.0_x64__qbz5n2kfra8p0\Lib\encodings\cp1252.py", line 19, in encode
return codecs.charmap_encode(input,self.errors,encoding_table)[0]
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
UnicodeEncodeError: 'charmap' codec can't encode character '\x92' in position 101: character maps to <undefined>
(spacewalks) (spacewalks)This is not what we were expecting! The problem is character
encoding. ‘Standard’ Latin characters are encoded using ASCII, but
the expanded Unicode character set covers many more. In this case, the
data contains Unicode characters that are represented in the ASCII input
file with shortcuts (Â as \u00c2 and
’ as \u0092).
When we read the file, Python converts those into the Unicode
characters. Then by default Windows tries to write out
eva-data.csv using UTF-7. This saves space compared to the
standard UTF-8, but it doesn’t include all of the characters. It
automatically converts \u0092 into the shorter
\x92, then discovers that doesn’t exist in UTF-7.
The fact that different systems have different defaults, which can change or even break your code’s behaviour, shows why it is so important to make our code’s requirements explicit!
We can easily fix this by explicitly telling Python what encoding to use when reading and writing our files:
data_f = open('./eva-data.json', 'r', encoding='ascii')
data_t = open('./eva-data.csv','w', encoding='utf-8')Do not foget to commit any files that have been changed.
Now we have the code running in a virtual environment, in the next episode we will inspect it in more detail, to see if we can understand and improve it.
Further reading
We recommend the following resources for some additional reading on the topic of this episode:
Also check the full reference set for the course.
Key Points
- Virtual environments keep Python versions and dependencies required by different projects separate.
- A Python virtual environment is itself a directory structure.
- You can use
venvto create and manage Python virtual environments, andpipto install and manage external dependencies your code relies on. - By convention, you can save and export your Python virtual
environment in
requirements.txtfile in your project’s root directory, which can then be shared with collaborators/users and used to replicate your virtual environment elsewhere.
Content from Code readability
Last updated on 2025-07-01 | Edit this page
Overview
Questions
- Why does code readability matter?
- How can I organise my code to be more readable?
- What types of documentation can I include to improve the readability of my code?
Objectives
After completing this episode, participants should be able to:
- Organise code into reusable functions that achieve a singular purpose
- Choose function and variable names that help explain the purpose of the function or variable
- Write informative comments and docstrings to provide more detail about what the code is doing
In this episode, we will introduce the concept of readable code and consider how it can help create reusable scientific software and empower collaboration between researchers.
When someone writes code, they do so based on requirements that are likely to change in the future. Requirements change because software interacts with the real world, which is dynamic. When these requirements change, the developer (who is not necessarily the same person who wrote the original code) must implement the new requirements. They do this by reading the original code to understand the different abstractions, and identify what needs to change. Readable code facilitates the reading and understanding of the abstraction phases and, as a result, facilitates the evolution of the codebase. Readable code saves future developers’ time and effort.
In order to develop readable code, we should ask ourselves: “If I re-read this piece of code in fifteen days or one year, will I be able to understand what I have done and why?” Or even better: “If a new person who just joined the project reads my software, will they be able to understand what I have written here?”
We will now learn about a few software best practices we can follow to help create more readable code.
Place import statements at the top
Let’s have a look our code again - the first thing we may notice is
that our script currently places import statements throughout the code.
Conventionally, all import statements are placed at the top of the
script so that dependent libraries are clearly visible and not buried
inside the code (even though there are standard ways of describing
dependencies - e.g. using requirements.txt file). This will
help readability (accessibility) and reusability of our code.
Our code after the modification should look like the following.
PYTHON
import json
import csv
import datetime as dt
import matplotlib.pyplot as plt
# https://data.nasa.gov/resource/eva.json (with modifications)
data_f = open('./eva-data.json', 'r', encoding='ascii')
data_t = open('./eva-data.csv','w', encoding='utf-8')
g_file = './cumulative_eva_graph.png'
fieldnames = ("EVA #", "Country", "Crew ", "Vehicle", "Date", "Duration", "Purpose")
data=[]
for i in range(374):
line=data_f.readline()
print(line)
data.append(json.loads(line[1:-1]))
#data.pop(0)
## Comment out this bit if you don't want the spreadsheet
w=csv.writer(data_t)
time = []
date =[]
j=0
for i in data:
print(data[j])
# and this bit
w.writerow(data[j].values())
if 'duration' in data[j].keys():
tt=data[j]['duration']
if tt == '':
pass
else:
t=dt.datetime.strptime(tt,'%H:%M')
ttt = dt.timedelta(hours=t.hour, minutes=t.minute, seconds=t.second).total_seconds()/(60*60)
print(t,ttt)
time.append(ttt)
if 'date' in data[j].keys():
date.append(dt.datetime.strptime(data[j]['date'][0:10], '%Y-%m-%d'))
#date.append(data[j]['date'][0:10])
else:
time.pop(0)
j+=1
t=[0]
for i in time:
t.append(t[-1]+i)
date,time = zip(*sorted(zip(date, time)))
plt.plot(date,t[1:], 'ko-')
plt.xlabel('Year')
plt.ylabel('Total time spent in space to date (hours)')
plt.tight_layout()
plt.savefig(g_file)
plt.show()Let’s make sure we commit our changes.
Use meaningful variable names
Variables are the most common thing you will assign when coding, and it’s really important that it is clear what each variable means in order to understand what the code is doing. If you return to your code after a long time doing something else, or share your code with a colleague, it should be easy enough to understand what variables are involved in your code from their names. Therefore we need to give them clear names, but we also want to keep them concise so the code stays readable. There are no “hard and fast rules” here, and it’s often a case of using your best judgment.
Some useful tips for naming variables are:
- Short words are better than single character names. For example, if
we were creating a variable to store the speed to read a file,
s(for ‘speed’) is not descriptive enough butMBReadPerSecondAverageAfterLastFlushToLogis too long to read and prone to misspellings.ReadSpeed(orread_speed) would suffice. - If you are finding it difficult to come up with a variable name that is both short and descriptive, go with the short version and use an inline comment to describe it further (more on those in the next section). This guidance does not necessarily apply if your variable is a well-known constant in your domain - for example, c represents the speed of light in physics.
- Try to be descriptive where possible and avoid meaningless or funny
names like
foo,bar,var,thing, etc.
There are also some restrictions to consider when naming variables in Python:
- Only alphanumeric characters and underscores are permitted in variable names.
- You cannot begin your variable names with a numerical character as
this will raise a syntax error. Numerical characters can be included in
a variable name, just not as the first character. For example,
read_speed1is a valid variable name, but1read_speedisn’t. (This behaviour may be different for other programming languages.) - Variable names are case sensitive. So
speed_of_lightandSpeed_Of_Lightare not the same. - Programming languages often have global pre-built functions, such as
input, which you may accidentally overwrite if you assign a variable with the same name and no longer be able to access the originalinputfunction. In this case, opting for something likeinput_datawould be preferable. Note that this behaviour may be explicitly disallowed in other programming languages but is not in Python.
Give a descriptive name to a variable
Below we have a variable called var being set the value
of 9.81. var is not a very descriptive name here as it
doesn’t tell us what 9.81 means, yet it is a very common constant in
physics! Go online and find out which constant 9.81 relates to and
suggest a new name for this variable.
Hint: the units are metres per second squared!
\[ 9.81 m/s^2 \] is the gravitational force exerted by the Earth. It is often referred to as “little g” to distinguish it from “big G” which is the Gravitational Constant. A more descriptive name for this variable therefore might be:
Rename our variables to be more descriptive
Let’s apply this to eva_data_analysis.py.
-
Edit the code as follows to use descriptive variable names:
- Change data_f to input_file
- Change data_t to output_file
- Change g_file to graph_file
What other variable names in our code would benefit from renaming?
Commit your changes to your repository. Remember to use an informative commit message.
Updated code:
PYTHON
import json
import csv
import datetime as dt
import matplotlib.pyplot as plt
# https://data.nasa.gov/resource/eva.json (with modifications)
input_file = open('./eva-data.json', 'r', encoding='ascii')
output_file = open('./eva-data.csv', 'w', encoding='utf-8')
graph_file = './cumulative_eva_graph.png'
fieldnames = ("EVA #", "Country", "Crew ", "Vehicle", "Date", "Duration", "Purpose")
data=[]
for i in range(374):
line=input_file.readline()
print(line)
data.append(json.loads(line[1:-1]))
#data.pop(0)
## Comment out this bit if you don't want the spreadsheet
w=csv.writer(output_file)
time = []
date =[]
j=0
for i in data:
print(data[j])
# and this bit
w.writerow(data[j].values())
if 'duration' in data[j].keys():
tt=data[j]['duration']
if tt == '':
pass
else:
t=dt.datetime.strptime(tt,'%H:%M')
ttt = dt.timedelta(hours=t.hour, minutes=t.minute, seconds=t.second).total_seconds()/(60*60)
print(t,ttt)
time.append(ttt)
if 'date' in data[j].keys():
date.append(dt.datetime.strptime(data[j]['date'][0:10], '%Y-%m-%d'))
#date.append(data[j]['date'][0:10])
else:
time.pop(0)
j+=1
t=[0]
for i in time:
t.append(t[-1]+i)
date,time = zip(*sorted(zip(date, time)))
plt.plot(date,t[1:], 'ko-')
plt.xlabel('Year')
plt.ylabel('Total time spent in space to date (hours)')
plt.tight_layout()
plt.savefig(graph_file)
plt.show()We should also rename variables w, t,
tt and ttt to be more descriptive.
Commit changes:
Use standard libraries
Our script currently reads the data line-by-line from the JSON data file and uses custom code to manipulate the data. Variables of interest are stored in lists but there are more suitable data structures (e.g. data frames) to store data in our case. By choosing custom code over standard and well-tested libraries, we are making our code less readable and understandable and more error-prone.
The main functionality of our code can be rewritten as follows using
the Pandas library to load and manipulate the data in data
frames.
First, we need to install this dependency into our virtual environment (which should be active at this point).
The code should now look like:
PYTHON
import matplotlib.pyplot as plt
import pandas as pd
# Data source: https://data.nasa.gov/resource/eva.json (with modifications)
input_file = open('./eva-data.json', 'r', encoding='ascii')
output_file = open('./eva-data.csv', 'w', encoding='utf-8')
graph_file = './cumulative_eva_graph.png'
eva_df = pd.read_json(input_file, convert_dates=['date'])
eva_df['eva'] = eva_df['eva'].astype(float)
eva_df.dropna(axis=0, inplace=True)
eva_df.sort_values('date', inplace=True)
eva_df.to_csv(output_file, index=False)
eva_df['duration_hours'] = eva_df['duration'].str.split(":").apply(lambda x: int(x[0]) + int(x[1])/60)
eva_df['cumulative_time'] = eva_df['duration_hours'].cumsum()
plt.plot(eva_df['date'], eva_df['cumulative_time'], 'ko-')
plt.xlabel('Year')
plt.ylabel('Total time spent in space to date (hours)')
plt.tight_layout()
plt.savefig(graph_file)
plt.show()We should replace the existing code in our Python script
eva_data_analysis.py with the above code and commit the
changes. Remember to use an informative commit message.
BASH
(venv_spacewalks) $ git add eva_data_analysis.py
(venv_spacewalks) $ git commit -m "Refactor code to use standard libraries"Make sure to capture the changes to your virtual development environment too.
Use comments to explain functionality
Commenting is a very useful practice to help convey the context of the code. It can be helpful as a reminder for your future self or your collaborators as to why code is written in a certain way, how it is achieving a specific task, or the real-world implications of your code.
There are several ways to add comments to code:
- An inline comment is a comment on the same line as a code statement. Typically, it comes after the code statement and finishes when the line ends and is useful when you want to explain the code line in short. Inline comments in Python should be separated by at least two spaces from the statement; they start with a # followed by a single space, and have no end delimiter.
- A multi-line or block comment can
span multiple lines and has a start and end sequence. To comment out a
block of code in Python, you can either add a # at the beginning of each
line of the block or surround the entire block with three single
(
''') or double quotes (""").
PYTHON
x = 5 # In Python, inline comments begin with the `#` symbol and a single space.
'''
This is a multiline
comment
in Python.
'''Here are a few things to keep in mind when commenting your code:
- Focus on the why and the how of your code - avoid using comments to explain what your code does. If your code is too complex for other programmers to understand, consider rewriting it for clarity rather than adding comments to explain it.
- Make sure you are not reiterating something that your code already conveys on its own. Comments should not echo your code.
- Keep comments short and concise. Large blocks of text quickly become unreadable and difficult to maintain.
- Comments that contradict the code are worse than no comments. Always make a priority of keeping comments up-to-date when code changes.
Examples of unhelpful comments
PYTHON
statetax = 1.0625 # Assigns the float 1.0625 to the variable 'statetax'
citytax = 1.01 # Assigns the float 1.01 to the variable 'citytax'
specialtax = 1.01 # Assigns the float 1.01 to the variable 'specialtax'The comments in this code simply tell us what the code does, which is easy enough to figure out without the inline comments.
Examples of helpful comments
PYTHON
statetax = 1.0625 # State sales tax rate is 6.25% through Jan. 1
citytax = 1.01 # City sales tax rate is 1% through Jan. 1
specialtax = 1.01 # Special sales tax rate is 1% through Jan. 1In this case, it might not be immediately obvious what each variable represents, so the comments offer helpful, real-world context. The date in the comment also indicates when the code might need to be updated.
Add comments to our code
- Examine
eva_data_analysis.py. Add as many comments as you think is required to help yourself and others understand what that code is doing. - Commit your changes to your repository. Remember to use an informative commit message.
Some good comments may look like the example below.
PYTHON
import matplotlib.pyplot as plt
import pandas as pd
# https://data.nasa.gov/resource/eva.json (with modifications)
input_file = open('./eva-data.json', 'r', encoding='ascii')
output_file = open('./eva-data.csv', 'w', encoding='utf-8')
graph_file = './cumulative_eva_graph.png'
print("--START--")
print(f'Reading JSON file {input_file}')
# Read the data from a JSON file into a Pandas dataframe
eva_df = pd.read_json(input_file, convert_dates=['date'])
eva_df['eva'] = eva_df['eva'].astype(float)
# Clean the data by removing any incomplete rows and sort by date
eva_df.dropna(axis=0, inplace=True)
eva_df.sort_values('date', inplace=True)
print(f'Saving to CSV file {output_file}')
# Save dataframe to CSV file for later analysis
eva_df.to_csv(output_file, index=False)
print(f'Plotting cumulative spacewalk duration and saving to {graph_file}')
# Plot cumulative time spent in space over years
eva_df['duration_hours'] = eva_df['duration'].str.split(":").apply(lambda x: int(x[0]) + int(x[1])/60)
eva_df['cumulative_time'] = eva_df['duration_hours'].cumsum()
plt.plot(eva_df['date'], eva_df['cumulative_time'], 'ko-')
plt.xlabel('Year')
plt.ylabel('Total time spent in space to date (hours)')
plt.tight_layout()
plt.savefig(graph_file)
plt.show()
print("--END--")Commit changes:
Separate units of functionality
Functions are a fundamental concept in writing software and are one of the core ways you can organise your code to improve its readability. A function is an isolated section of code that performs a single, specific task that can be simple or complex. It can then be called multiple times with different inputs throughout a codebase, but its definition only needs to appear once.
Breaking up code into functions in this manner benefits readability since the smaller sections are easier to read and understand. Since functions can be reused, codebases naturally begin to follow the Don’t Repeat Yourself principle which prevents software from becoming overly long and confusing. The software also becomes easier to maintain because, if the code encapsulated in a function needs to change, it only needs updating in one place instead of many. As we will learn in a future episode, testing code also becomes simpler when code is written in functions. Each function can be individually checked to ensure it is doing what is intended, which improves confidence in the software as a whole.
Callout
Decomposing code into functions helps with reusability of blocks of code and eliminating repetition, but, equally importantly, it helps with code readability and testing.
Looking at our code, you may notice it contains different pieces of functionality:
- reading the data from a JSON file
- converting and saving the data in the CSV format
- processing/cleaning the data and preparing it for analysis
- data analysis and visualising the results
Let’s refactor our code so that reading the data in JSON format into
a dataframe (step 1.) and converting it and saving to the CSV format
(step 2.) are extracted into separate functions. Let’s name those
functions read_json_to_dataframe and
write_dataframe_to_csv respectively. The main part of the
script should then be simplified to invoke these new functions, while
the functions themselves contain the complexity of each of these two
steps. We will continue to work on steps 3. and 4. above later on.
After the initial refactoring, our code may look something like the following.
PYTHON
import matplotlib.pyplot as plt
import pandas as pd
def read_json_to_dataframe(input_file):
print(f'Reading JSON file {input_file}')
# Read the data from a JSON file into a Pandas dataframe
eva_df = pd.read_json(input_file, convert_dates=['date'])
eva_df['eva'] = eva_df['eva'].astype(float)
# Clean the data by removing any incomplete rows and sort by date
eva_df.dropna(axis=0, inplace=True)
eva_df.sort_values('date', inplace=True)
return eva_df
def write_dataframe_to_csv(df, output_file):
print(f'Saving to CSV file {output_file}')
# Save dataframe to CSV file for later analysis
df.to_csv(output_file, index=False)
# Main code
print("--START--")
input_file = open('./eva-data.json', 'r', encoding='ascii')
output_file = open('./eva-data.csv', 'w', encoding='utf-8')
graph_file = './cumulative_eva_graph.png'
# Read the data from JSON file
eva_data = read_json_to_dataframe(input_file)
# Convert and export data to CSV file
write_dataframe_to_csv(eva_data, output_file)
print(f'Plotting cumulative spacewalk duration and saving to {graph_file}')
# Plot cumulative time spent in space over years
eva_data['duration_hours'] = eva_data['duration'].str.split(":").apply(lambda x: int(x[0]) + int(x[1])/60)
eva_data['cumulative_time'] = eva_data['duration_hours'].cumsum()
plt.plot(eva_data['date'], eva_data['cumulative_time'], 'ko-')
plt.xlabel('Year')
plt.ylabel('Total time spent in space to date (hours)')
plt.tight_layout()
plt.savefig(graph_file)
plt.show()
print("--END--")We have chosen to create functions for reading in and writing out
data files since this is a very common task within research software.
While these functions do not contain that many lines of code due to
using the pandas in-built methods that do all the complex
data reading, converting and writing operations, it can be useful to
package these steps together into reusable functions if you need to read
in or write out a lot of similarly structured files and process them in
the same way.
Use docstrings to document functions
Now that we have written some functions, it is time to document them so that we can quickly recall (and others looking at our code in the future can understand) what the functions doe without having to read the code.
Docstrings are a specific type of documentation that are provided within functions and Python classes. A function docstring should explain what that particular code is doing, what parameters the function needs (inputs) and what form they should take, what the function outputs (you may see words like ‘returns’ or ‘yields’ here), and errors (if any) that might be raised.
Providing these docstrings helps improve code readability since it makes the function code more transparent and aids understanding. Particularly, docstrings that provide information on the input and output of functions makes it easier to reuse them in other parts of the code, without having to read the full function to understand what needs to be provided and what will be returned.
Python docstrings are defined by enclosing the text with 3 double
quotes ("""). This text is also indented to the same level
as the code defined beneath it, which is 4 whitespaces by
convention.
Example of a multi-line docstring
PYTHON
def divide(x, y):
"""
Divide number x by number y.
Args:
x: A number to be divided.
y: A number to divide by.
Returns:
float: The division of x by y.
Raises:
ZeroDivisionError: Cannot divide by zero.
"""
return x / ySome projects may have their own guidelines on how to write docstrings, such as numpy. If you are contributing code to a wider project or community, try to follow the guidelines and standards they provide for code style.
As your code grows and becomes more complex, the docstrings can form the content of a reference guide allowing developers to quickly look up how to use the APIs, functions, and classes defined in your codebase. Hence, it is common to find tools that will automatically extract docstrings from your code and generate a website where people can learn about your code without downloading/installing and reading the code files - such as MkDocs.
Let’s write a docstring for the function
read_json_to_dataframe we introduced in the previous
exercise using the Google
Style Python Docstrings Convention. Remember, questions we want to
answer when writing the docstring include:
- What the function does?
- What kind of inputs does the function take? Are they required or optional? Do they have default values?
- What output will the function produce?
- What exceptions/errors, if any, it can produce?
Our read_json_to_dataframe function fully described by a
docstring may look like:
PYTHON
def read_json_to_dataframe(input_file):
"""
Read the data from a JSON file into a Pandas dataframe.
Clean the data by removing any incomplete rows and sort by date
Args:
input_file (str): The path to the JSON file.
Returns:
eva_df (pd.DataFrame): The cleaned and sorted data as a dataframe structure
"""
print(f'Reading JSON file {input_file}')
# Read the data from a JSON file into a Pandas dataframe
eva_df = pd.read_json(input_file, convert_dates=['date'])
eva_df['eva'] = eva_df['eva'].astype(float)
# Clean the data by removing any incomplete rows and sort by date
eva_df.dropna(axis=0, inplace=True)
eva_df.sort_values('date', inplace=True)
return eva_dfWriting docstrings
Write a docstring for the function
write_dataframe_to_csv we introduced earlier.
Our write_dataframe_to_csv function fully described by a
docstring may look like:
PYTHON
def write_dataframe_to_csv(df, output_file):
"""
Write the dataframe to a CSV file.
Args:
df (pd.DataFrame): The input dataframe.
output_file (str): The path to the output CSV file.
Returns:
None
"""
print(f'Saving to CSV file {output_file}')
# Save dataframe to CSV file for later analysis
df.to_csv(output_file, index=False)Finally, our code may look something like the following:
PYTHON
import matplotlib.pyplot as plt
import pandas as pd
def read_json_to_dataframe(input_file):
"""
Read the data from a JSON file into a Pandas dataframe.
Clean the data by removing any incomplete rows and sort by date
Args:
input_file (str): The path to the JSON file.
Returns:
eva_df (pd.DataFrame): The cleaned and sorted data as a dataframe structure
"""
print(f'Reading JSON file {input_file}')
# Read the data from a JSON file into a Pandas dataframe
eva_df = pd.read_json(input_file, convert_dates=['date'])
eva_df['eva'] = eva_df['eva'].astype(float)
# Clean the data by removing any incomplete rows and sort by date
eva_df.dropna(axis=0, inplace=True)
eva_df.sort_values('date', inplace=True)
return eva_df
def write_dataframe_to_csv(df, output_file):
"""
Write the dataframe to a CSV file.
Args:
df (pd.DataFrame): The input dataframe.
output_file (str): The path to the output CSV file.
Returns:
None
"""
print(f'Saving to CSV file {output_file}')
# Save dataframe to CSV file for later analysis
df.to_csv(output_file, index=False)
# Main code
print("--START--")
input_file = open('./eva-data.json', 'r', encoding='ascii')
output_file = open('./eva-data.csv', 'w', encoding='utf-8')
graph_file = './cumulative_eva_graph.png'
# Read the data from JSON file
eva_data = read_json_to_dataframe(input_file)
# Convert and export data to CSV file
write_dataframe_to_csv(eva_data, output_file)
print(f'Plotting cumulative spacewalk duration and saving to {graph_file}')
# Plot cumulative time spent in space over years
eva_data['duration_hours'] = eva_data['duration'].str.split(":").apply(lambda x: int(x[0]) + int(x[1])/60)
eva_data['cumulative_time'] = eva_data['duration_hours'].cumsum()
plt.plot(eva_data['date'], eva_data['cumulative_time'], 'ko-')
plt.xlabel('Year')
plt.ylabel('Total time spent in space to date (hours)')
plt.tight_layout()
plt.savefig(graph_file)
plt.show()
print("--END--")Do not forget to commit any uncommitted changes you may have and then push your work to GitHub.
Further reading
We recommend the following resources for some additional reading on the topic of this episode:
- 7 tell-tale signs of unreadable code
- ‘Code Readability Matters’ from the Guardian’s engineering blog
- PEP 8 Style Guide for Python
- Coursera: Inline commenting in Python
- Introducing Functions from Introduction to Python
- W3Schools.com Python Functions
Also check the full reference set for the course.
Key Points
- Readable code is easier to understand, maintain, debug and extend (reuse) - saving time and effort.
- Choosing descriptive variable and function names will communicate their purpose more effectively.
- Using comments and docstrings to describe parts of the code will help transmit understanding and context.
- Use libraries or packages for common functionality to avoid duplication.
- Creating functions from the smallest, reusable units of code will make the code more readable and help. compartmentalise which parts of the code are doing what actions and isolate specific code sections for reuse.
Content from Code structure
Last updated on 2025-07-01 | Edit this page
Overview
Questions
- How can we best structure code?
- What is a common code structure (pattern) for creating software that can read input from command line?
- What are conventional places to store data, code, results, tests, auxiliary information and metadata within our software or research project?
Objectives
After completing this episode, participants should be able to:
- Structure code into smaller, reusable components with a single responsibility/functionality.
- Use the common code pattern for creating software that can read input from command line
- Follow best practices for organising software/research project directories for improved readability, accessibility and reproducibility.
In the previous episode we have seen some tools and practices that can help up improve readability of our code - including breaking our code into small, reusable functions that perform one specific task. We are going to explore a bit more how using common code structures can improve readability, accessibility and reusability of our code, and will expand these practices on our (research or code) projects as a whole.
Functions for modular and reusable code
As we have already seen in the previous episode - functions play a key role in creating modular and reusable code. We are going to carry on improving our code following these principles:
- Each function should have a single, clear responsibility. This makes functions easier to understand, test, and reuse.
- Write functions that can be easily combined or reused with other functions to build more complex functionality.
- Functions should accept parameters to allow flexibility and reusability in different contexts; avoid hard-coding values inside functions/code (e.g. data files to read from/write to) and pass them as arguments instead.
Bearing in mind the above principles, we can further simplify the
main part of our code by extracting the code to process, analyse our
data and plot a graph into a separate function
plot_cumulative_time_in_space.
We can further extract the code to convert the spacewalk duration
text into a number to allow for arithmetic calculations (into a separate
function text_to_duration) and the code to add this
numerical data as a new column in our dataset (into a separate function
add_duration_hours_variable).
The main part of our code then becomes much simpler and more readable, only containing the invocation of the following three functions:
PYTHON
...
eva_data = read_json_to_dataframe(input_file)
write_dataframe_to_csv(eva_data, output_file)
plot_cumulative_time_in_space(eva_data, graph_file)
...Remember to add docstrings and comments to the new functions to explain their functionalities.
Our new code (with the three new functions
plot_cumulative_time_in_space,
text_to_duration and
add_duration_hours_variable) may look like the
following.
PYTHON
import matplotlib.pyplot as plt
import pandas as pd
def read_json_to_dataframe(input_file):
"""
Read the data from a JSON file into a Pandas dataframe.
Clean the data by removing any incomplete rows and sort by date
Args:
input_file (str): The path to the JSON file.
Returns:
eva_df (pd.DataFrame): The cleaned and sorted data as a dataframe structure
"""
print(f'Reading JSON file {input_file}')
eva_df = pd.read_json(input_file, convert_dates=['date'])
eva_df['eva'] = eva_df['eva'].astype(float)
eva_df.dropna(axis=0, inplace=True)
eva_df.sort_values('date', inplace=True)
return eva_df
def write_dataframe_to_csv(df, output_file):
"""
Write the dataframe to a CSV file.
Args:
df (pd.DataFrame): The input dataframe.
output_file (str): The path to the output CSV file.
Returns:
None
"""
print(f'Saving to CSV file {output_file}')
df.to_csv(output_file, index=False)
def text_to_duration(duration):
"""
Convert a text format duration "HH:MM" to duration in hours
Args:
duration (str): The text format duration
Returns:
duration_hours (float): The duration in hours
"""
hours, minutes = duration.split(":")
duration_hours = int(hours) + int(minutes)/6 # there is an intentional bug on this line (should divide by 60 not 6)
return duration_hours
def add_duration_hours_variable(df):
"""
Add duration in hours (duration_hours) variable to the dataset
Args:
df (pd.DataFrame): The input dataframe.
Returns:
df_copy (pd.DataFrame): A copy of df_ with the new duration_hours variable added
"""
df_copy = df.copy()
df_copy["duration_hours"] = df_copy["duration"].apply(
text_to_duration
)
return df_copy
def plot_cumulative_time_in_space(df, graph_file):
"""
Plot the cumulative time spent in space over years
Convert the duration column from strings to number of hours
Calculate cumulative sum of durations
Generate a plot of cumulative time spent in space over years and
save it to the specified location
Args:
df (pd.DataFrame): The input dataframe.
graph_file (str): The path to the output graph file.
Returns:
None
"""
print(f'Plotting cumulative spacewalk duration and saving to {graph_file}')
df = add_duration_hours_variable(df)
df['cumulative_time'] = df['duration_hours'].cumsum()
plt.plot(df.date, df.cumulative_time, 'ko-')
plt.xlabel('Year')
plt.ylabel('Total time spent in space to date (hours)')
plt.tight_layout()
plt.savefig(graph_file)
plt.show()
# Main code
print("--START--")
input_file = open('./eva-data.json', 'r')
output_file = open('./eva-data.csv', 'w')
graph_file = './cumulative_eva_graph.png'
eva_data = read_json_to_dataframe(input_file)
write_dataframe_to_csv(eva_data, output_file)
plot_cumulative_time_in_space(eva_data, graph_file)
print("--END--")
As you may notice, the main part of our code has now been majorly simplified and is much easier to follow.
Command-line interface to code
A common way to structure code is to have a command-line interface to allow the passing of various parameters. For example, we can pass the input data file to be read and the output file to be written to as parameters to our script and avoid hard-coding them. This improves interoperability and reusability of our code as it can now be run over any data file of the same structure, invoked from the command line terminal and integrated into other scripts or workflows/pipelines. For example, another script can produce our input data and can be “chained” with our code in a more complex data analysis pipeline. Another use case would be invoking our script in a loop to automatically analyse a number of input data files (compare that to running the script manually over hundreds or thousands of files - which is slow, error-prone and requires manual intervention).
There is a common code structure (pattern) for writing code with a command-line interface in Python:
PYTHON
# import modules
def main(args):
# perform some actions
if __name__ == "__main__":
# perform some actions before main()
main(args)In this pattern the main actions performed by the script are
contained within the main function (which does not need to
be called main, but using this convention helps others in
understanding your code). The main function is then called
within the if statement
__name__ == "__main__", after some other actions have been
performed (usually the parsing of command-line arguments, which will be
explained below). __name__ is a special variable which is
set by the Python interpreter before the execution of any code in the
source file. What value is given by the interpreter to
__name__ is determined by the manner in which the script is
invoked.
If we run the source file directly using the Python interpreter, e.g.:
then the interpreter will assign the hard-coded string
"__main__" to the __name__ variable:
However, if your script is imported by another Python script, e.g. in order to reuse its functions, with:
then the Python interpreter will assign the name “eva_data_analysis”
from the import statement to the __name__ variable (note
that import statement matches our script’s name):
Because of this behaviour of the Python interpreter, we can put any
code that should only be executed when running the script directly
within the if __name__ == "__main__": structure, allowing
the rest of the code within the script to be safely imported by another
script if we so wish.
While it may not seem very useful to have your script importable by another script, there are a number of situations in which you would want to do this:
- for testing of your code, you can have your testing framework import
your script, and run special test functions which then call the
mainfunction directly; - where you want to not only be able to run your script from the command-line, but also provide a programmer-friendly application programming interface (API) for advanced users.
We will use the sys library to read the command line
arguments passed to our script and make them available in our code as a
list - remember to import this library first.
Our modified code will now look as follows.
PYTHON
import json
import csv
import datetime as dt
import matplotlib.pyplot as plt
import pandas as pd
import sys
def main(input_file, output_file, graph_file):
print("--START--")
eva_data = read_json_to_dataframe(input_file)
write_dataframe_to_csv(eva_data, output_file)
plot_cumulative_time_in_space(eva_data, graph_file)
print("--END--")
def read_json_to_dataframe(input_file):
"""
Read the data from a JSON file into a Pandas dataframe.
Clean the data by removing any incomplete rows and sort by date
Args:
input_file (str): The path to the JSON file.
Returns:
eva_df (pd.DataFrame): The cleaned and sorted data as a dataframe structure
"""
print(f'Reading JSON file {input_file}')
eva_df = pd.read_json(input_file, convert_dates=['date'])
eva_df['eva'] = eva_df['eva'].astype(float)
eva_df.dropna(axis=0, inplace=True)
eva_df.sort_values('date', inplace=True)
return eva_df
def write_dataframe_to_csv(df, output_file):
"""
Write the dataframe to a CSV file.
Args:
df (pd.DataFrame): The input dataframe.
output_file (str): The path to the output CSV file.
Returns:
None
"""
print(f'Saving to CSV file {output_file}')
df.to_csv(output_file, index=False)
def text_to_duration(duration):
"""
Convert a text format duration "HH:MM" to duration in hours
Args:
duration (str): The text format duration
Returns:
duration_hours (float): The duration in hours
"""
hours, minutes = duration.split(":")
duration_hours = int(hours) + int(minutes)/6 # there is an intentional bug on this line (should divide by 60 not 6)
return duration_hours
def add_duration_hours_variable(df):
"""
Add duration in hours (duration_hours) variable to the dataset
Args:
df (pd.DataFrame): The input dataframe.
Returns:
df_copy (pd.DataFrame): A copy of df_ with the new duration_hours variable added
"""
df_copy = df.copy()
df_copy["duration_hours"] = df_copy["duration"].apply(
text_to_duration
)
return df_copy
def plot_cumulative_time_in_space(df, graph_file):
"""
Plot the cumulative time spent in space over years
Convert the duration column from strings to number of hours
Calculate cumulative sum of durations
Generate a plot of cumulative time spent in space over years and
save it to the specified location
Args:
df (pd.DataFrame): The input dataframe.
graph_file (str): The path to the output graph file.
Returns:
None
"""
print(f'Plotting cumulative spacewalk duration and saving to {graph_file}')
df = add_duration_hours_variable(df)
df['cumulative_time'] = df['duration_hours'].cumsum()
plt.plot(df.date, df.cumulative_time, 'ko-')
plt.xlabel('Year')
plt.ylabel('Total time spent in space to date (hours)')
plt.tight_layout()
plt.savefig(graph_file)
plt.show()
if __name__ == "__main__":
if len(sys.argv) < 3:
input_file = './eva-data.json'
output_file = './eva-data.csv'
print(f'Using default input and output filenames')
else:
input_file = sys.argv[1]
output_file = sys.argv[2]
print('Using custom input and output filenames')
graph_file = './cumulative_eva_graph.png'
main(input_file, output_file, graph_file)We can now run our script from the command line passing the JSON input data file and CSV output data file as:
Remember to commit our changes.
Directory structure for software projects
Expanding on the code structure theme, following conventions on consistent and informative directory structure for your projects will ensure people will immediately know where to find things within your project, especially helpful for long-term research projects or when working in teams. The directory structure for organising your research software project (or research projects in general) involves creating a clear and logical layout for files and data, ensuring easy navigation, collaboration and reproducibility.
Below are some good practices for setting up and maintaining a research project directory structure.
- Top-level directory
- Put all files related to a project into a single directory
- Choose a meaningful name that reflects the project’s purpose or topic.
- Subdirectories - organise the project into clear, well-labeled
sub-directories based on the type of content. Common categories include:
- Data - store raw, intermediate, and processed data in separate sub-directories to maintain clarity and avoid overwriting and losing your raw data
- Code/scripts/src - for storing your source code
- Results - for storing analysis outputs, summary statistics, or any data generated after processing.
- Documentation - include a detailed project description and documentation on how the project is organised, methodologies, and file dependencies.
- Figures/Plots - store all visualisations like charts, graphs, and figures generated from the analysis (these can also go in the results directory).
- References - a folder for research papers, articles, or any other literature cited or referenced in the project.
- Naming conventions
- Avoid special characters or spaces (they can cause errors when read by computers); use underscores (_) or hyphens (-) instead
- Name files to reflect their contents, version, or date (or use version control to track different versions).
- Use version control
- Code and data should be version controlled; you can also version control manuscripts, results, etc.
- If data files are too large (or contain sensitive information) to
track by version control, untrack them using
.gitignore - Use tags/releases to mark specific versions of results (a version submitted to a journal, dissertation version, poster version, etc.) so as to avoid using version numbers in file names and proliferation of different files.
OUTPUT
project_name/
├── README.md # overview of the project
├── data/ # data files used in the project
│ ├── README.md # describes where data came from
│ ├── raw/
│ └── processed/
├── manuscript/ # manuscript describing the results
├── results/ # results of the analysis (data, tables)
│ ├── preliminary/
│ └── final/
├── figures/ # results of the analysis (figures)
│ ├── comparison_plot.png
│ └── regression_chart.pdf
├── src/ # contains source code for the project
│ ├── LICENSE # license for your code
│ ├── requirements.txt # software requirements and dependencies
│ ├── main_script.py # main script/code entry point
│ └── ...
├── doc/ # documentation for your project
├── index.rst # entry point into the documentation website
└── ...Challenge
Refactor your software project so that input data is stored in
data/ directory and results (the graph and CSV data files)
saved in results/ directory. Remember to create the
results/ directory or your code will fail.
PYTHON
import matplotlib.pyplot as plt
import pandas as pd
import sys
# https://data.nasa.gov/resource/eva.json (with modifications)
def main(input_file, output_file, graph_file):
print("--START--")
eva_data = read_json_to_dataframe(input_file)
write_dataframe_to_csv(eva_data, output_file)
plot_cumulative_time_in_space(eva_data, graph_file)
print("--END--")
def read_json_to_dataframe(input_file):
"""
Read the data from a JSON file into a Pandas dataframe.
Clean the data by removing any incomplete rows and sort by date
Args:
input_file (str): The path to the JSON file.
Returns:
eva_df (pd.DataFrame): The cleaned and sorted data as a dataframe structure
"""
print(f'Reading JSON file {input_file}')
eva_df = pd.read_json(input_file, convert_dates=['date'])
eva_df['eva'] = eva_df['eva'].astype(float)
eva_df.dropna(axis=0, inplace=True)
eva_df.sort_values('date', inplace=True)
return eva_df
def write_dataframe_to_csv(df, output_file):
"""
Write the dataframe to a CSV file.
Args:
df (pd.DataFrame): The input dataframe.
output_file (str): The path to the output CSV file.
Returns:
(None):
"""
print(f'Saving to CSV file {output_file}')
df.to_csv(output_file, index=False)
def text_to_duration(duration):
"""
Convert a text format duration "HH:MM" to duration in hours
Args:
duration (str): The text format duration
Returns:
duration_hours (float): The duration in hours
"""
hours, minutes = duration.split(":")
duration_hours = int(hours) + int(minutes)/6 # there is an intentional bug on this line (should divide by 60 not 6)
return duration_hours
def add_duration_hours_variable(df):
"""
Add duration in hours (duration_hours) variable to the dataset
Args:
df (pd.DataFrame): The input dataframe.
Returns:
df_copy (pd.DataFrame): A copy of df_ with the new duration_hours variable added
"""
df_copy = df.copy()
df_copy["duration_hours"] = df_copy["duration"].apply(
text_to_duration
)
return df_copy
def plot_cumulative_time_in_space(df, graph_file):
"""
Plot the cumulative time spent in space over years
Convert the duration column from strings to number of hours
Calculate cumulative sum of durations
Generate a plot of cumulative time spent in space over years and
save it to the specified location
Args:
df (pd.DataFrame): The input dataframe.
graph_file (str): The path to the output graph file.
Returns:
(None):
"""
print(f'Plotting cumulative spacewalk duration and saving to {graph_file}')
df = add_duration_hours_variable(df)
df['cumulative_time'] = df['duration_hours'].cumsum()
plt.plot(df.date, df.cumulative_time, 'ko-')
plt.xlabel('Year')
plt.ylabel('Total time spent in space to date (hours)')
plt.tight_layout()
plt.savefig(graph_file)
plt.show()
if __name__ == "__main__":
if len(sys.argv) < 3:
input_file = 'data/eva-data.json'
output_file = 'results/eva-data.csv'
print(f'Using default input and output filenames')
else:
input_file = sys.argv[1]
output_file = sys.argv[2]
print('Using custom input and output filenames')
graph_file = 'results/cumulative_eva_graph.png'
main(input_file, output_file, graph_file)Remember to commit your latest changes:
Further reading
We recommend the following resources for some additional reading on the topic of this episode:
- Organizing your projects chapter from the CodeRefinery’s Reproducible Research tutorial
- MIT Broad Research Communication Lab’s “File Structure” guide
Also check the full reference set for the course.
Key Points
- Good practices for code and project structure are essential for creating readable, accessible and reproducibile projects.
Content from Code correctness
Last updated on 2025-06-17 | Edit this page
Overview
Questions
- How can we verify that our code is correct?
- How can we automate our software tests?
- What makes a “good” test?
- Which parts of our code should we prioritise for testing?
Objectives
After completing this episode, participants should be able to:
- Explain why code testing is important and how this supports FAIR software.
- Describe the different types of software tests (unit tests, integration tests, regression tests).
- Implement unit tests to verify that function behave as expected
using the Python testing framework
pytest. - Interpret the output from
pytestto identify which functions are not behaving as expected. - Write tests using typical values, edge cases and invalid inputs to ensure that the code can handle extreme values and invalid inputs appropriately.
- Evaluate code coverage to identify how much of the codebase is being tested and identify areas that need further tests.
Now that we have improved the structure and readability of our code - it is much easier to test its functionality and improve it further. The goal of software testing is to check that the actual results produced by a piece of code meet our expectations, i.e. are correct.
Why use software testing?
Adopting software testing as part of our research workflow helps us to conduct better research and produce FAIR software:
- Software testing can help us be more productive as it helps us to identify and fix problems with our code early and quickly and allows us to demonstrate to ourselves and others that our code does what we claim. More importantly, we can share our tests alongside our code, allowing others to verify our software for themselves.
- The act of writing tests encourages to structure our code as individual functions and often results in a more readable, modular and maintainable codebase that is easier to extend or repurpose.
- Software testing improves the accessibility and reusability of our code - well-written software tests capture the expected behaviour of our code and can be used alongside documentation to help other developers quickly make sense of our code. In addition, a well tested codebase allows developers to experiment with new features safe in the knowledge that tests will reveal if their changes have broken any existing functionality.
- Software testing underpins the FAIR process by giving us the confidence to engage in open research practices - if we are not sure that our code works as intended and produces accurate results, we are unlikely to feel confident about sharing our code with others. Software testing brings piece of mind by providing a step-by-step approach that we can apply to verify that our code is correct.
Types of software tests
There are many different types of software tests, including:
Unit tests focus on testing individual functions in isolation. They ensure that each small part of the software performs as intended. By verifying the correctness of these individual units, we can catch errors early in the development process.
Integration tests check how different parts of the code e.g. functions work together.
Regression tests are used to ensure that new changes or updates to the codebase do not adversely affect the existing functionality. They involve checking whether a program or part of a program still generates the same results after changes have been made.
End-to-end tests are a special type of integration testing which checks that a program as a whole behaves as expected.
In this course, our primary focus will be on unit testing. However, the concepts and techniques we cover will provide a solid foundation applicable to other types of testing.
Types of software tests
Fill in the blanks in the sentences below:
- __________ tests compare the ______ output of a program to its ________ output to demonstrate correctness.
- Unit tests compare the actual output of a ______ ________ to the expected output to demonstrate correctness.
- __________ tests check that results have not changed since the previous test run.
- __________ tests check that two or more parts of a program are working together correctly.
- End-to-end tests compare the actual output of a program to the expected output to demonstrate correctness.
- Unit tests compare the actual output of a single function to the expected output to demonstrate correctness.
- Regression tests check that results have not changed since the previous test run.
- Integration tests check that two or more parts of a program are working together correctly.
Informal testing
How should we test our code? One approach is to copy/paste the code
or a function into a Python terminal (different from a command line
terminal), which allows you to interact with the Python interpreter more
directly.
From the Python terminal we can then run one function or a piece of code
at a time and check that they behave as expected. As input to our
code/function we are testing, we typically use some input values for
which we know what the correct return value should be.
Let’s do this for our text_to_duration function. Recall
that the text_to_duration function converts a spacewalk
duration stored as a string in format “HH:MM” to a duration in hours -
e.g. duration 01:15 (1 hour and 15 minutes) should return a
numerical value of 1.25.
PYTHON
def text_to_duration(duration):
"""
Convert a text format duration "HH:MM" to duration in hours
Args:
duration (str): The text format duration
Returns:
duration_hours (float): The duration in hours
"""
hours, minutes = duration.split(":")
duration_hours = int(hours) + int(minutes)/6
return duration_hoursTo start a Python terminal, you simply type python3
(with no other parameters) from the root directory of your project in a
command line terminal.
This will open an interactive Python terminal for you, which may look like this:
PYTHON
(venv_spacewalks) mbassan2@C-U-LOSXQ677L astronaut-data-analysis % python3
Python 3.11.7 (main, Dec 4 2023, 18:10:11) [Clang 15.0.0 (clang-1500.1.0.2.5)] on darwin
Type "help", "copyright", "credits" or "license" for more information.
>>> Once inside the Python terminal, you can start typing Python code.
The Python terminal will interactively run your code and return and
print results. We could copy and paste the code of our
text_to_duration function, but it is much simpler and more
elegant to import and then invoke it.
So, we have invoked our function with the value “10:00” and it returned the floating point value “10.0” as expected.
We can then further explore the behaviour of our function by running:
This all seems correct so far.
Testing code in this “informal” way in an important process to go through as we draft our code for the first time. Another tool that can help here is the Jupyter Notebook - like the Python terminal, the Jupyter Notebook is an interactive environment for writing and running code. It is a GUI tool which supports all kinds of interactive outputs, including many interactive visualisation libraries.
However, there are some serious drawbacks to this approach if used as our only form of testing.
What are the limitations of informally testing code? (5 minutes)
Think about the questions below. Your instructors may ask you to share your answers in a shared notes document and/or discuss them with other participants.
- Why might we choose to test our code informally?
- What are the limitations of relying solely on informal tests to verify that a piece of code is behaving as expected?
It can be tempting to test our code informally because this approach:
- is quick and easy
- provides immediate feedback
However, there are limitations to this approach:
- Working interactively is error prone
- We must reload our function in Python terminal each time we change our code
- We must repeat our tests every time we update our code which is time consuming
- We must rely on memory to keep track of how we have tested our code, e.g. what input values we tried
- We must rely on memory to keep track of which functions have been tested and which have not (informal testing may work well on smaller pieces of code but it becomes unpractical for a large codebase)
- Once we close the Python terminal, we lose all the test scenarios we have tried
Formal testing
We can overcome some of these limitations by formalising our testing process. A formal approach to testing our code is to write dedicated test functions to check it. These test functions:
- Run the function we want to test - the target function with known inputs
- Compare the output to known, valid results
- Raise an error if the function’s actual output does not match the expected output
- Are recorded in a test script that can be re-run on demand.
Let’s explore this process by writing some formal tests for our
text_to_duration function.
In VS Code, create a new Python file test_code.py in the
root of our project directory to store our tests.
Like before in the Python terminal, we need to import
text_to_duration into our test script. Then, we add our
first test function:
PYTHON
from eva_data_analysis import text_to_duration
def test_text_to_duration_integer():
input_value = "10:00"
test_result = text_to_duration("10:00") == 10
print(f"text_to_duration('10:00') == 10? {test_result}")
test_text_to_duration_integer()We can run this code from the command line terminal as:
This test checks that when we apply text_to_duration to
input value 10:00, the output matches the expected value of
10.
In this example, we use a print statement to report whether the
actual output from text_to_duration meets our
expectations.
However, this does not meet our requirement to “Raise an error if the function’s output does not match the expected output” and means that we must carefully read our test function’s output to identify whether it has failed.
To ensure that our code raises an error if the function’s output does
not match the expected output, we use Python’s assert
statement. The assert statement in Python checks whether a
condition is True or False. If the statement
is True, assert does not return a value and
the code continues to run. However, if the statement is
False, assert raises an
AssertError.
Let’s rewrite our test with an assert statement:
PYTHON
from eva_data_analysis import text_to_duration
def test_text_to_duration_integer():
assert text_to_duration("10:00") == 10
test_text_to_duration_integer()Notice that when we run test_text_to_duration_integer(),
nothing happens - there is no output. That is because our function is
working correctly and returning the expected value of 10.
Let’s add another test to check what happens when duration is not an integer number and if our function can handle durations with a non-zero minute component, and rerun our test code.
PYTHON
from eva_data_analysis import text_to_duration
def test_text_to_duration_float():
assert text_to_duration("10:15") == 10.25
def test_text_to_duration_integer():
assert text_to_duration("10:00") == 10
test_text_to_duration_float()
test_text_to_duration_integer()ERROR
(venv_spacewalks) $ python3 test_code.py
Traceback (most recent call last):
File "/Users/user/Desktop/spacewalks/test_code.py", line 9, in <module>
test_text_to_duration_float()
File "/Users/user/Desktop/spacewalks/test_code.py", line 4, in test_text_to_duration_float
assert text_to_duration("10:15") == 10.25
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
AssertionErrorNotice that this time, our test
test_text_to_duration_float fails. Our assert statement has
raised an AssertionError - a clear signal that there is a
problem in our code that we need to fix.
We know that duration 10:15 should be converted to
number 10.25. What is wrong with our code? If we look at
our text_to_duration function, we may identify the
following line of our code as problematic:
You may notice that our conversion code is wrong - the minutes component should have been divided by 60 and not 6. We were able to spot this tiny bug only by testing our code (note that just by looking at the result graph there is not way to spot incorrect results).
Let’s fix the problematic line and rerun out tests.
This time our tests run without problem.
Should we add more tests or the tests we have so far are enough? What
happens if our duration value is 10:20 (ten hours and 20
minutes) and our result is not a rational floating point number (like
10.25) but an irrational number such as
10.333333333? Let’s tests for this.
PYTHON
from eva_data_analysis import text_to_duration
def test_text_to_duration_float():
assert text_to_duration("10:20") == 10.333333
def test_text_to_duration_integer():
assert text_to_duration("10:00") == 10
test_text_to_duration_float()
test_text_to_duration_integer()ERROR
(venv_spacewalks) $ python3 test_code.py
Traceback (most recent call last):
File "/Users/user/work/SSI/lessons/astronaut-data-analysis/test_code.py", line 17, in <module>
test_text_to_duration_float()
File "/Users/user/work/SSI/lessons/astronaut-data-analysis/test_code.py", line 9, in test_text_to_duration_float
assert text_to_duration("10:20") == 10.333333
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
AssertionErrorOn computer systems, representation of irrational numbers is
typically not exact as they do not have an exact binary representation.
For this reason, we cannot use a simple double equals sign
(==) to compare the equality of floating point numbers.
Instead, we check that our floating point numbers are equal within a
very small tolerance (e.g. 1e-5). Hence, our code should look like:
PYTHON
...
def test_text_to_duration_float():
assert abs(text_to_duration("10:20") - 10.33333333) < 1e-5
...You may have noticed that we have to repeat a lot of code to add each individual test for each test case. You may also have noticed that our test script stopped after the first test failure and none of the tests after that were run. To run our remaining tests we would have to manually comment out our failing test and re-run the test script. As our code base grows, testing in this way becomes cumbersome and error-prone. These limitations can be overcome by automating our tests using a testing framework.
Testing frameworks
Testing frameworks can automatically find all the tests in our code base, run all of them (so we do not have to invoke them explicitly or, even worse, forget to invoke them), and present the test results as a readable summary.
We will use the Python testing framework pytest with its
code coverage extension pytest-cov. To install these
libraries into our virtual environment, from the command line terminal
do:
Let’s make sure that our tests are ready to work with
pytest.
-
pytestautomatically discovers tests based on specific naming patterns. It looks for files that start with “test_” or end with “_test.py”. Then, within these files,pytestlooks for functions that start with “test_”. Our test file already meets these requirements, so there is nothing to do here. However, our script does contain lines to run each of our test functions. These are no-longer required as pytest will run our tests so we can remove them: It is also conventional when working with a testing framework to place test files in a
testsdirectory at the root of our project and to name each test file after the code file that it targets. This helps in maintaining a clean structure and makes it easier for others to understand where the tests are located.
A set of tests for a given piece of code is called a test suite. Our
test suite is currently located in the root folder of our project. Let’s
move it to a dedicated test folder and rename our
test_code.py file to test_eva_analysis.py.
You can do it from VS Code or by typing the following commands in the command line terminal:
Before we re-run our tests using pytest, let’s update
our second test to use pytest’s function
approx() which is specifically intended for comparing
floating point numbers within a tolerance.
PYTHON
import pytest
from eva_data_analysis import text_to_duration
def test_text_to_duration_float():
assert text_to_duration("10:20") == pytest.approx(10.33333333)
def test_text_to_duration_integer():
assert text_to_duration("10:00") == 10Let’s also add some inline comments to clarify what each test is doing and expand our syntax to highlight the logic behind our approach:
PYTHON
import pytest
from eva_data_analysis import text_to_duration
def test_text_to_duration_float():
"""
Test that text_to_duration returns expected ground truth values
for typical durations with a non-zero minute component
"""
actual_result = text_to_duration("10:20")
expected_result = 10.33333333
assert actual_result == pytest.approx(expected_result)
def test_text_to_duration_integer():
"""
Test that text_to_duration returns expected ground truth values
for typical whole hour durations
"""
actual_result = text_to_duration("10:00")
expected_result = 10
assert actual_result == expected_result
Writing our tests this way highlights the key idea that each test should compare the actual results returned by our function with expected values.
Similarly, writing inline comments for our tests that complete the sentence “Test that …” helps us to understand what each test is doing and why it is needed.
Before running out tests with pytest, let’s reintroduce
our old bug in function text_to_duration that affects the
durations with a non-zero minute component like “10:20” but not those
that are whole hours, e.g. “10:00”:
Finally, let’s run our tests with pytest from our
project’s root directory (and not tests directory):
ERROR
========================================== test session starts ===========================================
platform darwin -- Python 3.11.7, pytest-8.3.3, pluggy-1.5.0
rootdir: /Users/user/work/SSI/lessons/astronaut-data-analysis-not-so-good
plugins: cov-5.0.0
collected 2 items
tests/test_code.py F. [100%]
================================================ FAILURES ================================================
________________________________________ test_text_to_duration_float _____________________________________
def test_text_to_duration_float():
> assert text_to_duration("10:20") == pytest.approx(10.33333333)
E assert 13.333333333333334 == 10.33333333 ± 1.0e-05
E
E comparison failed
E Obtained: 13.333333333333334
E Expected: 10.33333333 ± 1.0e-05
tests/test_code.py:5: AssertionError
=========================================== short test summary info =======================================
FAILED tests/test_code.py::test_text_to_duration_float - assert 13.333333333333334 == 10.33333333 ± 1.0e-05
========================================= 1 failed, 1 passed in 0.67s =====================================
From the above output from pytest’s execution of out
tests, we notice that:
- If a test function finishes without triggering an assertion, the
test is considered successful and is marked with a dot
(
.). - If an assertion fails or an error occurs, the test is marked as a
failure with an
F, and the output includes details about the error to help identify what went wrong.
Let’s fix our bug once again, and rerun our tests using
pytest.
OUTPUT
========================================== test session starts ===========================================
platform darwin -- Python 3.11.7, pytest-8.3.3, pluggy-1.5.0
rootdir: /Users/user/work/SSI/lessons/astronaut-data-analysis-not-so-good
plugins: cov-5.0.0
collected 2 items
tests/test_code.py .. [100%]
=========================================== 2 passed in 0.63s =============================================This time, all out tests passed.
Interpreting pytest output
A colleague has asked you to conduct a pre-publication review of their code which analyses time spent in space by various individual astronauts.
You tested their code using pytest, and got the
following output. Inspect it and answer the questions below.
Example pytest output
OUTPUT
============================================================ test session starts
platform darwin -- Python 3.12.3, pytest-8.2.2, pluggy-1.5.0
rootdir: /Users/Desktop/AnneResearcher/projects/Spacetravel
collected 9 items
tests/test_analyse.py FF.... [ 66%]
tests/test_prepare.py s.. [100%]
====================================================================== FAILURES
____________________________________________________________ test_total_duration
def test_total_duration():
durations = [10, 15, 20, 5]
expected = 50/60
actual = calculate_total_duration(durations)
> assert actual == pytest.approx(expected)
E assert 8.333333333333334 == 0.8333333333333334 ± 8.3e-07
E
E comparison failed
E Obtained: 8.333333333333334
E Expected: 0.8333333333333334 ± 8.3e-07
tests/test_analyse.py:9: AssertionError
______________________________________________________________________________ test_mean_duration
def test_mean_duration():
durations = [10, 15, 20, 5]
expected = 12.5/60
> actual = calculate_mean_duration(durations)
tests/test_analyse.py:15:
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
durations = [10, 15, 20, 5]
def calculate_mean_duration(durations):
"""
Calculate the mean of a list of durations.
"""
total_duration = sum(durations)/60
> mean_duration = total_duration / length(durations)
E NameError: name 'length' is not defined
Spacetravel.py:45: NameError
=========================================================================== short test summary info
FAILED tests/test_analyse.py::test_total_duration - assert 8.333333333333334 == 0.8333333333333334 ± 8.3e-07
FAILED tests/test_analyse.py::test_mean_duration - NameError: name 'length' is not defined
============================================================== 2 failed, 6 passed, 1 skipped in 0.02s - How many tests has our colleague included in the test suite?
- The first test in test_prepare.py has a status of s; what does this mean?
- How many tests failed?
- Why did “test_total_duration” fail?
- Why did “test_mean_duration” fail?
- 9 tests were detected in the test suite
- s - stands for “skipped”,
- 2 tests failed: the first and second tests in test file
test_analyse.py -
test_total_durationfailed because the calculated total duration differs from the expected value by a factor of 10 i.e. the assertionactual == pytest.approx(expected)evaluated toFalse -
test_mean_durationfailed because there is a syntax error incalculate_mean_duration. Our colleague has used the commandlength(not a python command) instead oflen. As a result, running the function returns aNameErrorrather than a calculated value and the test assertion evaluates toFalse.
Test suite design
We now have the tools in place to automatically run tests. However, that alone is not enough to properly test code. We will now look into what makes a good test suite and good practices for testing code.
Let’s start by considering the following scenario. A collaborator on
our project has sent us the following code which adds a new column
called crew_size to our data containing the number of
astronauts participating in any given spacewalk. How do we know that it
works as intended and that it will not break the rest of our code? For
this, we need to write a test suite with a comprehensive coverage of the
new code.
PYTHON
import matplotlib.pyplot as plt
import pandas as pd
import sys
import re # added this line
# https://data.nasa.gov/resource/eva.json (with modifications)
def main(input_file, output_file, graph_file):
print("--START--")
eva_data = read_json_to_dataframe(input_file)
eva_data = add_crew_size_column(eva_data) # added this line
write_dataframe_to_csv(eva_data, output_file)
plot_cumulative_time_in_space(eva_data, graph_file)
print("--END--")
...
def calculate_crew_size(crew):
"""
Calculate the size of the crew for a single crew entry
Args:
crew (str): The text entry in the crew column containing a list of crew member names
Returns:
(int): The crew size
"""
if crew.split() == []:
return None
else:
return len(re.split(r';', crew))-1
def add_crew_size_column(df):
"""
Add crew_size column to the dataset containing the value of the crew size
Args:
df (pd.DataFrame): The input data frame.
Returns:
df_copy (pd.DataFrame): A copy of df with the new crew_size variable added
"""
print('Adding crew size variable (crew_size) to dataset')
df_copy = df.copy()
df_copy["crew_size"] = df_copy["crew"].apply(
calculate_crew_size
)
return df_copy
...
Writing good tests
The aim of writing good tests is to verify that each of our functions behaves as expected with the full range of inputs that it might encounter. It is helpful to consider each argument of a function in turn and identify the range of typical values it can take. Once we have identified this typical range or ranges (where a function takes more than one argument), we should:
- Test all values at the edge of the range
- Test at least one interior point
- Test invalid values
Let’s have a look at the calculate_crew_size function
from our colleague’s new code and write some tests for it.
Unit tests for calculate_crew_size
Implement unit tests for the calculate_crew_size
function. Cover typical cases and edge cases.
Hint - use the following template when writing tests:
def test_MYFUNCTION (): # FIXME
"""
Test that ... #FIXME
"""
# Typical value 1
actual_result = _______________ #FIXME
expected_result = ______________ #FIXME
assert actual_result == expected_result
# Typical value 2
actual_result = _______________ #FIXME
expected_result = ______________ #FIXME
assert actual_result == expected_result
We can add the following test functions to out test suite.
PYTHON
import pytest
from eva_data_analysis import (
text_to_duration,
calculate_crew_size
)
...
def test_calculate_crew_size():
"""
Test that calculate_crew_size returns expected ground truth values
for typical crew values
"""
actual_result = calculate_crew_size("Valentina Tereshkova;")
expected_result = 1
assert actual_result == expected_result
actual_result = calculate_crew_size("Judith Resnik; Sally Ride;")
expected_result = 2
assert actual_result == expected_result
# Edge cases
def test_calculate_crew_size_edge_cases():
"""
Test that calculate_crew_size returns expected ground truth values
for edge case where crew is an empty string
"""
actual_result = calculate_crew_size("")
assert actual_result is NoneLet’s run out tests:
Parameterising tests
Looking at out new test functions, we may notice that they do not follow the “Don’t Repeat Yourself principle” which prevents software - including testing code - from becoming repetitive and too long. In our test code, a small block of code is repeated twice with different input values:
PYTHON
def test_calculate_crew_size():
"""
Test that calculate_crew_size returns expected ground truth values
for typical crew values
"""
actual_result = calculate_crew_size("Valentina Tereshkova;")
expected_result = 1
assert actual_result == expected_result
actual_result = calculate_crew_size("Judith Resnik; Sally Ride;")
expected_result = 2
assert actual_result == expected_resultTo avoid such repetitions in our test code, we use test
parameterisation. This allows us to apply our test function to
a list of input/expected output pairs without the need for repetition.
To parameterise the test_calculate_crew_size function, we
can rewrite is as follows:
PYTHON
@pytest.mark.parametrize("input_value, expected_result", [
("Valentina Tereshkova;", 1),
("Judith Resnik; Sally Ride;", 2),
])
def test_calculate_crew_size(input_value, expected_result):
"""
Test that calculate_crew_size returns expected ground truth values
for typical crew values
"""
actual_result = calculate_crew_size(input_value)
assert actual_result == expected_resultNotice the following key changes to our code:
- The unparameterised version of
test_calculate_crew_sizefunction did not have any arguments and our input/expected values were all defined in the body of our test function. - In the parameterised version of
test_calculate_crew_size, the body of our test function has been rewritten as a parameterised block of code that uses the variablesinput_valueandexpected_resultwhich are now arguments of the test function. - A Python decorator
@pytest.mark.parametrizeis placed immediately before the test function and indicates that it should be run once for each set of parameters provided.
Callout
In Python, a decorator is a function that can modify the behaviour of
another function. @pytest.mark.parametrize is a decorator
provided by pytest that modifies the behaviour of our test
function by running it multiple times - once for each set of inputs.
This decorator takes two main arguments:
Parameter names: a string with the names of the parameters that the test function will accept, separated by commas – in this case “input_value” and “expected_value”
Parameter values: a list of tuples, where each tuple contains the values for the parameters specified in the first argument.
As you can see, the parameterised version of our test function is shorter, more readable and easier to maintain.
Just enough tests
In this episode, so far we have (only) written tests for two
individual functions: text_to_duration and
calculate_crew_size.
We can quantify the proportion of our code base that is run (also referred to as “exercised”) by a given test suite using a metric called code coverage:
\[ \text{Line Coverage} = \left( \frac{\text{Number of Executed Lines}}{\text{Total Number of Executable Lines}} \right) \times 100 \]
We can calculate our test coverage using the pytest-cov
library as follows.
OUTPUT
========================================================== test session starts
platform darwin -- Python 3.12.3, pytest-8.2.2, pluggy-1.5.0
rootdir: /Users/AnnResearcher/Desktop/Spacewalks
plugins: cov-5.0.0
collected 4 items
tests/test_eva_data_analysis.py .... [100%]
---------- coverage: platform darwin, python 3.12.3-final-0 ----------
Name Stmts Miss Cover
-----------------------------------------------------
eva_data_analysis.py 56 38 32%
tests/test_eva_data_analysis.py 20 0 100%
-----------------------------------------------------
TOTAL 76 38 50%
=========================================================== 4 passed in 1.04sTo get an in-depth report about which parts of our code are tested
and which are not, we can add the option
--cov-report=html.
This option generates a folder htmlcov in the project
root directory containing a code coverage report in HTML format. This
provides structured information about our test coverage including:
- a table showing the proportion of lines in each function that are currently tested, and
- an annotated copy of our code where untested lines are highlighted in red.
Ideally, all the lines of code in our code base should be exercised by at least one test. However, if we lack the time and resources to test every line of our code we should:
- avoid testing Python’s built-in functions or functions imported from
well-known and well-tested libraries like
pandasornumpy. - focus on the the parts of our code that carry the greatest “reputational risk”, i.e. that could affect the accuracy of our reported results.
Callout
Test coverage of less than 100% indicates that more testing may be helpful.
Test coverage of 100% does not mean that our code is bug-free.
Evaluating code coverage
Generate the code coverage report for your software using the
python3 -m pytest --cov --cov-report=html command.
Inspect the htmlcov folder created by the above command
in the root directory of your propject, then open the
htmlcov/index.html file in a Web browser and extract the
following information:
- What proportion of the code base is currently “not” exercised by the test suite?
- Which functions in our code base are currently untested?
- You can find this information on the “Files” tab of the HTML report
saved in the
htmlcov/index.htmlfile. The proportion of the code base NOT covered by our tests is 68% (100% - 32%) - this may differ for your version of the code. - You can find this information on the “Functions” tab of the HTML
report. The following functions in our code base are currently untested:
- read_json_to_dataframe
- write_dataframe_to_csv
- add_duration_hours_variable
- plot_cumulative_time_in_space
- add_crew_size_variable
At this point, now is a good time to commit our test suite to our codebase and push the changes to GitHub.
BASH
(venv_spacewalks) $ git add eva_data_analysis.py
(venv_spacewalks) $ git commit -m "Add additional analysis functions"
(venv_spacewalks) $ git add tests/
(venv_spacewalks) $ git commit -m "Add test suite"
(venv_spacewalks) $ python3 -m pip freeze > requirements.txt
(venv_spacewalks) $ git add requirements.txt
(venv_spacewalks) $ git commit -m "Added pytest and pytest-cov libraries."
(venv_spacewalks) $ git push origin main(Optional) More practice with a test suite
There is an optional exercise to implement additional tests and practice writing tests some more.
Continuous Integration for automated testing
Continuous Integration (CI) services provide the infrastructure to automatically run every test function in the test code suite every time changes are pushed to a remote repository. There is an extra episode on configuring CI for automated tests on GitHub for some additional reading.
Summary
During this episode, we have covered how to use software tests to
verify the correctness of our code. We have seen how to write a unit
test, how to manage and run our tests using the pytest
framework and how identify which parts of our code require additional
testing using test coverage reports.
These skills reduce the probability that there will be a mistake in our code and support reproducible research by giving us the confidence to engage in open research practices. Tests also document the intended behaviour of our code for other developers and mean that we can experiment with changes to our code knowing that our tests will let us know if we break any existing functionality. In other words, software testing supports the FAIR software principles by making our code more accessible and reusable.
Further reading
We recommend the following resources for some additional reading on the topic of this episode:
- The Defensive Programming episode from the Software Carpentry Python Programming lesson
- Using Python to double check your work (Software Sustainability Blog Post) by Peter Inglesby
- The Python Testing and Continuous Integration lesson on The Carpentries Incubator by François Michonneau
- [Test Driven Development][york-tdd-blog (University of York Research Coding Club Blog Post) by Peter Hill and Stephen Biggs-Fox
- Automated testing - Preventing yourself and others from breaking your functioning code Coderefinery lesson
- The Automatically Testing Software episode from the Intermediate Research Software Development lesson on The Carpentries Incubator by Aleksandra Nenadic et al.
Also check the full reference set for the course.
Key Points
- Code testing supports the FAIR principles by improving the accessibility and re-usability of research code.
- Unit testing is crucial as it ensures each functions works correctly.
- Using the
pytestframework, you can write basic unit tests for Python functions to verify their correctness. - Identifying and handling edge cases in unit tests is essential to ensure your code performs correctly under a variety of conditions.
- Test coverage can help you to identify parts of your code that require additional testing.
Content from Software documentation
Last updated on 2025-06-17 | Edit this page
Overview
Questions
- How should we document our code?
- Why are documentation and repository metadata important and how they support FAIR software?
- What are the minimum elements of documentation needed to support FAIR software?
Objectives
After completing this episode, participants should be able to:
- Use a
READMEfile to provide an overview and aCITATION.cfffile to add citation instructions to a code repository - Describe the main types of software documentation (tutorials, how to guides, reference and explanation).
- Apply a documentation framework to write effective documentation of any type.
- Describe the different formats available for delivering software documentation (Markdown files, wikis, static webpages).
- Implement MkDocs to generate and manage comprehensive project documentation
We have seen how writing inline comments and docstrings within our code can help with improving its readability. The purpose of software documentation is to communicate other important information about our software (its purpose, dependencies, how to install and run it, etc.) to the people who need it – both users and developers.
Activate your virtual environment
Why document our software?
Software documentation is often perceived as a thankless and time-consuming task with few tangible benefits and is often neglected in research projects. However, like software testing, documenting our software can help us and others conduct better research and produce FAIR software:
- Good documentation captures important methodological details ready for when we come to publish our research
- Good documentation can help us return to a project seamlessly after time away
- Documentation can facilitate collaborations by helping us onboard new project members quickly and more easily
- Good documentation can save us time by answering frequently asked questions (FAQs) about our code for us
- Software documentation supports the FAIR research software
principles by improving the re-usability of our code.
- Good documentation can make our software more understandable and reusable by others, and can bring us some citations and credit
- How-to guides and tutorials ensure that users can install our software independently and make use of its basic features
- Reference guides and background information can help developers understand our code sufficiently to modify/extend/repurpose it.
Code-level documentation
In previous episodes we encountered several different forms of in-code documentation aspects, including in-line comments and docstrings. These are an excellent way to improve the readability of our code, but by themselves are insufficient to ensure that our code is easy to use, understand and modify - this requires additional software-level documentation.
There are many different types of software-level documentation.
Technical documentation
Software-level technical documentation encompasses:
- Tutorials - lessons that guide learners through a series of
exercises to build proficiency as using the code
- How-To Guides - step by step instructions on how to accomplish specific goals using the code.
- Reference - a lookup manual to help users find relevant information about the software e.g. functions and their parameters.
- Explanation - conceptual discussion of the code to help users understand implementation decisions
Project-level documentation
Project-level documentation includes various information and metadata about software that help to discover it, explain the legal terms of reusing it, describe its functionality on a high level and how to install, run and contribute to it.
Repository metadata files
A common way to to provide project-level documentation is to include various metadata files in the software repository together with code. Many of these files can be described as “social documentation”, i.e. they indicate how users should “behave” in relation to our software project. Some common examples of repository metadata files and their role are explained below:
| File | Description |
|---|---|
| README | Provides an overview of the project, including installation, usage instructions, dependencies and links to other metadata files and technical documentation (tutorial/how-to/explanation/reference) |
| CONTRIBUTING | Explains to developers how to contribute code to the project including processes and standards that should be followed |
| CODE_OF_CONDUCT | Defines expected standards of conduct when engaging in a software project |
| LICENSE | Defines the (legal) terms of using, modifying and distributing the code |
| CITATION | Provides instructions on how to cite the code |
| AUTHORS | Provides information on who authored the code (can also be included in CITATION) |
Just enough documentation
For many small projects the following three pieces of project-level documentation may be sufficient: README, LICENSE and CITATION.
Let’s look at each of these files in turn.
README file
A README file acts as a “landing page” for your code repository on GitHub and should provide sufficient information for users to and developers to get started using your code.
README and the FAIR principles
Think about the question below. Your instructors may ask you to share your answer in a shared notes document and/or discuss them with other participants.
Here are some of the major sections you might find in a typical README. Which are essential to support the FAIR principles? Which are optional?
- Purpose of the code
- Audience (who the code is intended for)
- Installation instructions
- Contribution guide
- How to get help
- License
- Software citation
- Usage example
- Dependencies and their versions
- FAQs
- Code of Conduct
To support the FAIR principles (Findability, Accessibility, Interoperability, and Reusability), certain sections in a README file are more important than others. Below is a breakdown of the sections that are essential or optional in a README to align with these principles.
Essential
- Purpose of the code - clearly explains what the code does; essential for findability and reusability.
- Installation instructions - provides step-by-step instructions on how to install the software, ensuring accessibility.
- Usage Example - provides examples of how to use the code, helping users understand its functionality and enhancing reusability.
- License- specifies the terms under which the code can be used, which is crucial for legal clarity and reusability.
- Dependencies and their versions - lists the external libraries and tools required to run the code, including their versions; essential for reproducibility and interoperability.
- Software citation - provides citation information for academic use, ensuring proper attribution and reusability.
Optional
- Audience (who the code is intended for) - helps users identify if the code is relevant to them, improving findability and usability.
- How to get help - informs users where they can get help, ensuring better accessibility.
- Contribution guide - encourages and guides contributions from the community, enhancing the code’s development and reusability.
- FAQs - provide answers to common questions, aiding in troubleshooting and improving accessibility.
- Code of Conduct - sets expectations for behaviour in the community, fostering a welcoming environment and enhancing accessibility.
Let’s create a simple README for our repository - from VS Code or
command line terminal create file README.md (in Markdown
format) or README.txt (in plain text format).
We can start by adding a one-liner that explains the purpose of our code and who it is for.
# Spacewalks
## Overview
Spacewalks is a Python analysis tool for researchers to generate visualisations
and statistical summaries of NASA's extravehicular activity datasets.Now let’s add a list of Spacewalks’ key features:
## Features
Key features of Spacewalks:
- Generates a CSV table of summary statistics of extravehicular activity crew sizes
- Generates a line plot to show the cumulative duration of space walks over timeNow let’s tell users about any pre-requisites required to run the software:
## Pre-requisites
Spacewalks was developed using Python version 3.12
To install and run Spacewalks you will need have Python >=3.12
installed. You will also need the following libraries (minimum versions in brackets)
- [NumPy](https://www.numpy.org/) >=2.0.0 - Spacewalk's test suite uses NumPy's statistical functions
- [Matplotlib](https://matplotlib.org/stable/index.html) >=3.0.0 - Spacewalks uses Matplotlib to make plots
- [pytest](https://docs.pytest.org/en/8.2.x/#) >=8.2.0 - Spacewalks uses pytest for testing
- [pandas](https://pandas.pydata.org/) >= 2.2.0 - Spacewalks uses pandas for data frame manipulation Spacewalks README
Extend the README for Spacewalks by adding:
- Installation instructions
- A simple usage example
Installation instructions:
NB: In the solution below the back ticks of each code block have been escaped to avoid rendering issues (if you are copying and pasting the text, make sure you unescape them).
## Installation instructions
+ Clone the Spacewalks repository to your local machine using Git.
If you don't have Git installed, you can download it from the official Git website.
\`\`\`
git clone https://github.com/your-repository-url/spacewalks.git
cd spacewalks
\`\`\`
+ Install the necessary dependencies:
\`\`\`
python3 -m pip install pandas==2.2.2 matplotlib==3.8.4 numpy==2.0.0 pytest==7.4.2
\`\`\`
+ To ensure everything is working correctly, run the tests using pytest.
\`\`\`
python3 -m pytest
\`\`\`Usage instructions:
## Usage Example
To run an analysis using the eva_data_analysis.py script from the command line terminal,
launch the script using Python as follows:
\`\`\`
# Usage Examples
python3 eva_data_analysis.py eva-data.json eva-data.csv
\`\`\`
The first argument is path to the JSON data file.
The second argument is the path the CSV output file.LICENSE file
Copyright allows a creator of work (such as written text, photographs, films, music, software code) to state that they own the work they have created. Copyright is automatically implied - even if the creator does not explicitly assert it, copyright of the work exists from the moment of creation. A licence is a legal document which sets down the terms under which the creator is releasing what they have created for others to use, modify, extend or exploit.
Because any creative work is copyrighted the moment it is created, even without any kind of licence agreement, it is important to state the terms under which software can be reused. The lack of a licence for your software implies that no one can reuse the software at all - hence it is imperative you declare it. A common way to declare your copyright of a piece of software and the license you are distributing it under is to include a file called LICENSE in the root directory of your code repository.
There is an optional extra episode in this course on different open source software licences that you can choose for your code and that we recommend for further reading.
Tools to help you choose a licence
- A short intro on different open source software licences included as extra content to this course.
- The open source guide on applying, changing and editing licenses.
- choosealicense.com online tool has some great resources to help you choose a license that is appropriate for your needs, and can even automate adding the LICENSE file to your GitHub code repository.
Select a licence
Choose a license for your code. Discuss with your neighbour or the group your choice of license and reason for choosing it.
Add a license to your code
Add a LICENSE file containing the full text of your chosen license to your code repository.
- Licence can be done in either of the following two ways:
- Create a LICENSE file in the root of your software repository on your local machine and copy into it the text of your chosen licence (you can find it online). Push your local changes to your GitHub repository.
- In your repository on GitHub, go to
Add fileoption and start typing file name “LICENSE” - GitHub will recognise that you want to add a licence and will offer you a choice of difference licences to choose from. Select one and commit your changes, then dogit pulllocally to bring those changes to your machine.
- Add a copyright statement, the name of the license you are using and
a mention of the LICENSE file to at least one source code file
(e.g.
eva_data_analysis.py) - Link to your LICENSE file from README to make this information about your code more prominent.
After completing the above, check the “About” section of your repository’s GitHub landing webpage and see if there is now a license listed.
CITATION file
We should add a citation file to our repository to provide instructions on how to cite our code. A citation file can be a plain text (CITATION.txt) or a Markdown file (CITATION.md), but there are certain benefits to using use a special file format called the Citation File Format (CFF), which provides a way to include richer metadata about code (or datasets) we want to cite, making it easy for both humans and machines to use this information.
Why use CFF?
For developers, using a CFF file can help to automate the process of publishing new releases on Zenodo via GitHub. GitHub also “understands” CFF, and will display citation information prominently on the landing page of a repository that contains citation info in CFF.
For users, having a CFF file makes it easy to cite the software or dataset with formatted citation information available for copy+paste and direct import from GitHub into reference managers like Zotero.
CFF file format
A CFF file is using the YAML key-value pair format. At a minimum a CFF file must contain the title of the software/data, the type of asset (software or data) and at least one author:
YAML
# This CITATION.cff file was generated with cffinit.
# Visit https://bit.ly/cffinit to generate yours today!
cff-version: 1.2.0
title: My Software
message: >-
If you use this software, please cite it using the
metadata from this file.
type: software
authors:
- given-names: Anne
family-names: ResearcherAdditional and optional metadata includes an abstract, repository URL and more.
Creating CFF file and making your software citable
We can create (and later update) a CFF file for our software using an
online application called cffinit by following
these steps:
Head to
cffinitonline tool.-
Then, work through the metadata input form to complete the minimum information needed to generate a CFF.
We can use the following description as an “Abstract”: “A Python script to analyse NASA extravehicular activity data”
Add the URL of the code repository as a “Related Resources”.
Add at least two key words under the “Keywords” section.
At the end of the process, download the CFF file and inspect it. It should look like this:
YAML
# This CITATION.cff file was generated with cffinit.
# Visit https://bit.ly/cffinit to generate yours today!
cff-version: 1.2.0
title: Spacewalks
message: >-
If you use this software, please cite it using the
metadata from this file.
type: software
authors:
- given-names: Jaffa
name-particle: Sarah
- given-names: Aleksandra
family-names: Nenadic
- given-names: Kamilla
family-names: Kopec-Harding
repository-code: >-
https://github.com/YOUR-REPOSITORY-URL/spacewalks.git
abstract: >-
A Python script to analyse NASA extravehicular activity
data
keywords:
- NASA
- Extravehicular activityCFF files can also be updated using the cffinit online
tool by pasting the previous version of CFF file and working through the
form to update various fields.
Citing
To cite our software (or dataset), once a CFF file has been pushed to our remote repository, GitHub’s “Cite this repository” button can be used to generate a citation in various formats (APA, BibTeX).
Further information is available from the Turing Way’s guide to software citation.
Spacewalks software citation
Add the citation file for our Spacewalks software to the root folder of our repository on GitHub. You can either do it directly on GitHub or creating the file locally and the committing and pushing to GitHub from the command line.
Documentation tools
Once our project reaches a certain size or level of complexity we may want to add additional documentation such as a standalone tutorial or “background” explaining our methodological choices.
Once we move beyond using a README as our primary source of documentation, we need to consider how we will distribute our documentation to our users. Options include:
- A
docs/folder of Markdown files - Adding a Wiki to our repository
- Creating a set of web pages for our documentation using a static site generator for our documentation such as Sphinx or MkDocs.
Creating a static site is a popular solution as it has the key benefit being able to automatically generate a reference manual from any docstrings we have added to our code.
MkDocs
Let’s setup the static documentation site generator tool MkDocs.
BASH
python3 -m pip install mkdocs
python3 -m pip install "mkdocstrings[python]"
python3 -m pip install mkdocs-materialLet’s creates a new MkDocs project in the current directory:
OUTPUT
INFO - Writing config file: ./mkdocs.yml
INFO - Writing initial docs: ./docs/index.mdThis command creates a new MkDocs project in the current directory
with a docs folder containing an index.md file
and a mkdocs.yml configuration file in the root of our
project.
Now, let’s fill in the mkdocs.yml configuration file for
our project.
YAML
site_name: Spacewalks Documentation
use_directory_urls: false
theme:
name: "material"
font: false
nav:
- Spacewalks Documentation: index.md
- Tutorials: tutorials.md
- How-To Guides: how-to-guides.md
- Reference: reference.md
- Background: explanation.mdNote font: false variable is for GDPR compliance;
use_directory_url: false variable tells MKDocs tools how to
handle URLs for documentation that is served as a website (we will cover
this in a moment).
Let’s add support for mkdocstrings - this will allow us
to automatically add our docstrings into our documentation using a
simple tag.
YAML
site_name: Spacewalks Documentation
use_directory_urls: false
theme:
name: "material"
font: false
nav:
- Spacewalks Documentation: index.md
- Tutorials: tutorials.md
- How-To Guides: how-to-guides.md
- Reference: reference.md
- Background: explanation.md
plugins:
- mkdocstringsLet’s populate our docs/ folder to match our
configuration file.
BASH
(venv_spacewalks) $ touch docs/tutorials.md
(venv_spacewalks) $ touch docs/how-to-guides.md
(venv_spacewalks) $ touch docs/reference.md
(venv_spacewalks) $ touch docs/explanation.mdLet’s populate our reference file reference.md with some
preamble to include before the reference manual that will be generated
from the docstrings we created.
MARKDOWN
This file documents the key functions in the Spacewalks tool.
It is provided as a reference manual.
::: eva_data_analysisFinally, let’s build our documentation.
OUTPUT
INFO - Cleaning site directory
INFO - Building documentation to directory: /Users/AnnResearcher/Desktop/Spacewalks/site
WARNING - griffe: eva_data_analysis.py:105: No type or annotation for returned value 'int'
WARNING - griffe: eva_data_analysis.py:84: No type or annotation for returned value 1
WARNING - griffe: eva_data_analysis.py:33: No type or annotation for returned value 1
INFO - Documentation built in 0.31 secondsOnce the build step is completed, our documentation site is saved to
a site folder in the root of our project folder. These
files will be distributed with our code. We can either direct users to
read these files locally on their own device using their browser, or we
can choose to host our documentation as a website that our uses can
navigate to.
Note that we used the setting use_directory_urls: false
in the mkdocs.yml file. This setting ensures that the
documentation site is generated with URLs that are easy to navigate
locally on a user’s device.
Explore your documentation
Explore documentation in site/ folder built with MkDocs
for your project, starting from the index.html file.
Open index.html file in a Web browser to see how it
renders.
Check site/reference.html to see how docstrings from
your functions are provided here as a reference manual.
Finally, let us commit our documentation to the main branch of our git repository and push the changes to GitHub.
BASH
(venv_spacewalks) $ git add mkdocs.yml
(venv_spacewalks) $ git add docs/
(venv_spacewalks) $ git add site/
(venv_spacewalks) $ git commit -m "Add project-level documentation"
(venv_spacewalks) $ python3 -m pip freeze > requirements.txt
(venv_spacewalks) $ git add requirements.txt
(venv_spacewalks) $ git commit -m "Added mkdocs plugin"
(venv_spacewalks) $ git push origin mainHosting documentation
We saw how MkDocs documentation can be distributed with our repository and viewed “offline” using a browser.
We can also make our documentation available as a live website by deploying our documentation to a hosting service.
GitHub Pages
As our repository is hosted in GitHub, we can use GitHub Pages - a service that allows GitHub users to host websites directly from their GitHub repositories.
There are two types of GitHub Pages: project pages and user/organization Pages. While similar, they have different deployment workflows, and we will only discuss project pages here. For information about deploying to user/organisational pages, see MkDocs Deployment pages.
Project Pages deploy site files to a branch (by default the
gh-pages branch, but it can be any other branch) within the
project repository. To deploy our documentation:
Warning! Before we proceed to the next step, we MUST ensure that there are no uncommitted changes or untracked files in our repository.
If there are, the commands used in the upcoming steps will include them in our documentation!
- (If not done already), let us commit our documentation to the main branch of our git repository and push the changes to GitHub.
BASH
(venv_spacewalks) $ git add mkdocs.yml
(venv_spacewalks) $ git add docs/
(venv_spacewalks) $ git add site/
(venv_spacewalks) $ git commit -m "Add project-level documentation"
(venv_spacewalks) $ git push origin main- Once we are on the main branch and all our changes are up to date, run the following command from the command line termindal to deploy our documentation to GitHub.
BASH
# Important:
# - This command will push the documentation to the gh-pages branch of your repository
# - It will ALSO include uncommitted changes and untracked files (read the warning above!!) <- VERY IMPORTANT!!
(venv_spacewalks) $ mkdocs gh-deployOUTPUT
INFO - Cleaning site directory
INFO - Building documentation to directory: /Users/AnnResearch/Desktop/Spacewalks/site
WARNING - griffe: eva_data_analysis.py:105: No type or annotation for returned value 'int'
WARNING - griffe: eva_data_analysis.py:84: No type or annotation for returned value 1
WARNING - griffe: eva_data_analysis.py:33: No type or annotation for returned value 1
INFO - Documentation built in 0.37 seconds
WARNING - Version check skipped: No version specified in previous deployment.
INFO - Copying '/Users/AnnResearcher/Desktop/Spacewalks/site' to 'gh-pages' branch and pushing to
GitHub.
Enumerating objects: 63, done.
Counting objects: 100% (63/63), done.
Delta compression using up to 11 threads
Compressing objects: 100% (60/60), done.
Writing objects: 100% (63/63), 578.91 KiB | 7.93 MiB/s, done.
Total 63 (delta 7), reused 0 (delta 0), pack-reused 0
remote: Resolving deltas: 100% (7/7), done.
remote:
remote: Create a pull request for 'gh-pages' on GitHub by visiting:
remote: https://github.com/kkh451/spacewalks/pull/new/gh-pages
remote:
To https://github.com/kkh451/spacewalks-dev.git
* [new branch] gh-pages -> gh-pages
INFO - Your documentation should shortly be available at: https://kkh451.github.io/spacewalks/This command will build our documentation with MkDocs, then commit
and push the files to the gh-pages branch using the GitHub
ghp-import tool which is installed as a dependency of
MkDocs.
For more options, use:
Notice that the deploy command did not allow us to preview the site before it was pushed to GitHub; so, it is a good idea to check changes locally with the build commands before deploying.
Other options
You can find out about other deployment options including free documentation hosting service ReadTheDocs on the MkDocs deployment pages.
Documentation guides
Once we start to consider other forms of documentation beyond the README, we can also increase reusability of our code by ensuring that the content and style of our documentation matches its purpose.
Documentation guides such as Write the Docs, The Good Docs Project and the Diataxis framework provide a range of resources including documentation templates to help to help us do this.
Spacewalks how-to guide
Review the Diataxis guidance page on writing a How-to guide. Identify three features of an effective how-to guide.
Following the Diataxis guidelines, add a how-to guide to the
docs/how-to-guides.mdfile in your documentation folder to show users how to change the destination filename for the output CSV dataset generated by the Spacewalks software.
An effective how-to guide should:
- be goal oriented and focus on action.
- avoid teaching or explanation
- use appropriate language e.g. conditional imperatives
- have an informative title
An example how-to guide for our project to the file
docs/how-to-guides.md:
# How to change the file path of Spacewalk's output dataset
This guide shows you how to set the file path for Spacewalk's output
data set to a location of your choice.
By default, the cleaned data set in CSV format, generated by the Spacewalk software, is saved to the `results/`
folder within the working directory with file name `eva-data.csv`.
If you would like to modify the name or location of the output dataset, set the
second command line argument to your chosen file path.
For example, if you want to save the output data set to the subfolder `data/clean/` you can
invoke the script as:
`(venv_spacewalks) $ python3 eva_data_analysis.py eva-data.json data/clean/eva-data-clean.csv`
The specified destination folder `data/clean/` must exist before running spacewalks analysis script.Remember to rebuild your documentation:
The Diataxis framework provides guidance for developing technical documentation for different purposes. Tutorials differ in purpose and scope to how-to guides, and as a result, differ in content and style.
Spacewalks tutorial
Let’s adapt the how-to guide from the previous challenge into a tutorial that explains how to change the file path for the output dataset when running the analysis script.
Here is what an example tutorial may look like.
Introduction
In this tutorial, we will learn how to change the file path for the output dataset generated by the Spacewalk software. By the end of this tutorial, you will be able to specify a custom file path for the cleaned dataset.
Prerequisites
Before you start, ensure you have the following:
- Python installed on your system
- The Spacewalk script (
eva_data_analysis.py) - An input dataset (
eva-data.json)
Prepare the destination directory
First, let us decide where we want to save the cleaned dataset and make sure the directory exists.
For this tutorial, we will use data/clean/ as the
destination folder.
Let’s create the directory if it does not exist - e.g. from the command line do:
Run the analysis script with a custom path
Next, execute the Spacewalk script and specify the custom file path for the output dataset:
Replace data/eva-data.json) and data/clean/eva-data-clean.csv).
Here is the complete command:
BASH
(venv_spacewalks) $ python3 eva_data_analysis.py data/eva-data.json data/clean/eva-data-clean.csvNotice how the output to the command line clearly indicates that we are using a custom output file path.
OUTPUT
Using custom input and output filenames
Reading JSON file data/eva-data.json
Saving to CSV file data/clean/eva-data-clean.csv
Adding crew size variable (crew_size) to dataset
Saving to CSV file data/clean/eva-data-clean.csv
Plotting cumulative spacewalk duration and saving to results/cumulative_eva_graph.pngAfter running the script, let us check the data/clean
directory to ensure the cleaned dataset has been saved correctly.
You should see eva-data-clean.csv file listed in the
data/clean folder.
Exercise: custom output path
- Create a new directory named
output/datain your working directory. - Run the Spacewalk script to save the cleaned dataset in the newly
created
output/datadirectory with the filenamecleaned-eva-data.csv. - Verify that the dataset has been saved correctly.
Solution
BASH
# Create the directory:
(venv_spacewalks) $ mkdir -p output/data
# Run the script:
(venv_spacewalks) $ python3 eva_data_analysis.py data/eva-data.json output/data/cleaned-eva-data.csv
# Verify the output:
(venv_spacewalks) $ ls output/data
# You should see cleaned-eva-data.csv listedCongratulations! You have successfully changed the file path for Spacewalks output dataset and completed an exercise to practice the process. You can now customize the output location and filename according to your needs.
Now that we have seen examples of both a how-to guide and a tutorial, let’s compare the two.
Tutorial vs. how-to guide
How does the content and language of our example tutorial differ from our example how-to guide?
Do not forget to commit any uncommitted changes you may have and then push your work to GitHub.
Further reading
We recommend the following resources for some additional reading on the topic of this episode:
- “The Art of Readme” article by Kira Oakley - a useful discussion of best practices for writing a high-quality README
- What are best practices for research software documentation? (Software Sustainability blog post) by Stephan Druskat et al.
- Preparing Software for Reuse and Release episode from the Intermediate Research Software Development lesson on The Carpentries Incubator by Aleksandra Nenadic et al.
- CodeRefinery lesson: How to document your research software
- CITATION.cff file format
Also check the full reference set for the course.
Key Points
- Documentation allows users to run and understand software without having to work things out for themselves directly from the source code.
- Software documentation supports the FAIR principles by improving the reusability of research code.
- A (good) README, CITATION file and LICENSE file are the minimum documentation elements required to support FAIR research code.
- Documentation can be provided to users in a variety of formats
including a
docsfolder of Markdown files, a repository Wiki and static webpages. - A static documentation site can be created using the tool MkDocs.
- Documentation frameworks such as Diataxis provide content and style guidelines that can helps us write high quality documentation.
Content from Open software management & collaboration
Last updated on 2025-06-17 | Edit this page
Overview
Questions
- How do I ensure my code is citable?
- How do we track issues with code in GitHub?
- How can we ensure that multiple developers can work on the same files simultaneously?
Objectives
After completing this episode, participants should be able to:
- Understand how to archive code to Zenodo and create a digital object identifier (DOI) for a software project (and include that info in CITATION.cff).
- Understand how to track issues with code in GitHub.
- Understand how to use Git branches for working on code in parallel and how to merge code back using pull requests.
- Apply issue tracking, branching and pull requests together to fix bugs while allowing other developers to work on the same code.
Sharing code openly promotes collaboration, transparency, and innovation by allowing others to review, use, and improve the code. It fosters knowledge exchange, accelerates scientific progress, and enhances the reproducibility of research. Additionally, open sharing encourages community contributions and can lead to better-maintained, more reliable software.
Adding a license and other metadata to our code (covered in the previous episode) are the first steps towards sharing the code publicly. There are several other important steps to consider which we will cover here.
Sharing code to encourage collaboration
Making the code public
By default repositories created on GitHub are private and only their creator can see them. Since we added an open source license to our repository we probably want to make sure people can actually access it.
To make your repository public, if it is not already, go to your
repository on GitHub and click on the Settings link near
the top right corner. Then scroll down to the bottom of the page and the
“Danger Zone” settings. Click on “Change Visibility” and you should see
a message saying “Change to public”. If it says “Change to private” then
the repository is already public. You will then be asked to confirm that
you indeed want to make the repository public and agree to the warning
that the code will now be publicly visible. As a security measure, you
will then have to put in your GitHub password.
Transferring to an organisation
Currently our repository is under the GitHub “namespace” of our individual user. This is OK for individual projects where we are the sole or at least the main code author, but for bigger and more complex projects it is common to use a GitHub organisation named after our project. If we are a member of an organisation and have the appropriate permissions then we can transfer a repository from our personal namespace to the organisation’s. This can be done with another option in the “Danger Zone” settings, the “Transfer ownership” button. Pressing this will then prompt us as to which organisation we want to transfer the repository to.
Archiving code to Zenodo and obtaining a DOI
Zenodo is a data archive run by CERN. Anybody can upload datasets up to 50GB to it and receive a Digital Object Identifier (DOI). Zenodo’s definition of a dataset is quite broad and can include code - which gives us a way to obtain a DOI for our software.
Let us now look into how we can archive a GitHub repository to Zenodo. Note that, instead of using the real Zenodo website, we will practice with Zenodo Sandbox.
Zenodo Sandbox
Zenodo Sandbox is a testing environment for Zenodo, a repository for research outputs, allowing users to safely experiment with its features without affecting the live system. It is a clone of Zenodo, created for testing purposes, that works exactly the same way as Zenodo you can use it for learning, training, experimenting, and preparing uploads without impacting the primary Zenodo repository until you are ready to publish and release your code (or other research outputs) officially. It will also not create real DOIs for a number of test repositories we use for this course and saturate the DOI space (remember that a DOI, once created, is meant to exist forever).
We can archive our GitHub repository to Zenodo (Sandbox) by doing the following:
- Go to the Zenodo Sandbox login page and choose to login with GitHub.
- Authorise Zenodo Sandbox to connect to GitHub.
- Go to the GitHub page in your Zenodo Sandbox account. This can be found in the pull down menu with your user name in the top right corner of the screen.
- You will now have a list of all of your GitHub repositories. Next to each will be an “On” button. If you have created a new repository you might need to press the “Sync” button to update the list of repositories Zenodo Sandbox knows about.
- Press the “On” button for the repository you want to archive. If this was successful you will be told to refresh the page.
- The repository should now appear in the list of “Enabled” repositories at the top of the screen, but it does not yet have a DOI. To get one we have to make a “release” on GitHub. Click on the repository and then press the green button to create a release. This will take you to GitHub’s release page where you will be asked to give a title and description of the release. You will also have to create a “tag” for your release - a way of having a friendly name for the version of some code in Git instead of using a long hash code. Often we will create a sequential version number for each release of the software and have the tag name match this, for example v1.0 or just 1.0.
- If we now refresh the Zenodo Sandbox page for this repository we will see that it has been assigned a DOI.
The DOI does not just link to GitHub, Zenodo will have taken a copy
(a snapshot) of our repository at the point where we tagged the release.
This means that even if we delete it from GitHub or even if GitHub were
ever to go away or remove it, there will still be a copy on Zenodo.
Zenodo will allow people to download the entire repository (more
accurately, its state at the time it was tagged for release) as a single
zip file.
Zenodo will have actually created two DOIs for you. One represents the latest version of the software and will always represent the latest if you make more releases. The other is specific to the release you made and will always point to that version. We can see both of these by clicking on the DOI link in the Zenodo page for the repository.
One of the things which is displayed on this page is a badge image that you can copy the link for and add to the README file in your GitHub repository so that people can find the Zenodo version of the repository. If you click on the DOI image in the Details section of the Zenodo page then you will be shown instructions for obtaining a link to the DOI badge in various formats including Markdown. Here is the badge for this repository and the corresponding Markdown:
[](https://doi.org/10.5281/zenodo.11869450)Archive your repository to Zenodo (Sandbox)
Note: for this exercise, as demonstrated earlier, you should use the Sandbox Zenodo (a version of Zenodo for testing and playing with before minting a real DOI). For real software releases, you should use Zenodo.
- Create an account on Zenodo Sandbox that is linked to your GitHub account.
- Use Zenodo Sandbox to create a release for your repository and obtain a DOI for it.
- Get the link to the DOI badge for your repository and add a link to this image to your README file in Markdown format. Check that this is the DOI for the latest version and not the DOI for a specific version, if not you will be updating this every time you make a release.
Problems with GitHub and Zenodo integration
The integration between GitHub and Zenodo does not interact well with
some browser privacy features and extensions. Firefox can be
particularly problematic with this and might open new tabs to login to
GitHub and then give an error saying:
Your browser did something unexpected. Please try again. If the error continues, try disabling all browser extensions.
If this happens try disabling the extra privacy features/extensions or
using another browser such as Chrome.
Adding a DOI and ORCID to the citation file
Now that we have our DOI it is good practice to include this
information in our citation file. Earlier we created a
CITATION.cff file with information about how to cite our
code. There are a few fields we can add now which are related to the
DOI; one of these is the version file which covers the
version number of the software. We can add a DOI to the file in the
identifiers section with a type of doi and
value of the Zenodo URL. Optionally we can also add a
date-released field indicating the date we released this
software. Here is an updated version of our CITATION.cff
from the previous episode with a version number, DOI and release date
added.
YAML
# This CITATION.cff file was generated with cffinit.
# Visit https://bit.ly/cffinit to generate yours today!
cff-version: 1.2.0
title: Spacewalks
message: >-
If you use this software, please cite it using the
metadata from this file.
type: software
authors:
- given-names: Jaffa
name-particle: Sarah
- given-names: Aleksandra
family-names: Nenadic
- given-names: Kamilla
family-names: Kopec-Harding
repository-code: >-
https://github.com/YOUR-REPOSITORY-URL/spacewalks.git
abstract: >-
A Python script to analyse NASA extravehicular activity
data
keywords:
- NASA
- Extravehicular activity
version: 1.0.1
identifiers:
- type: doi
value: 10.5281/zenodo.1234
date-released: 2024-06-01Add a DOI to your citation file
Add the DOI you were allocated in the previous exercise to your
CITATION.cff file and then commit and push the updated
version to your GitHub repository. If you used the commit
field in your CITATION.cff file before to point to a given
version of the code - you can now remove it as using the DOI field is
better for this job.
Going further with publishing code
We now have our code published online, licensed as open source, archived with Zenodo, accessible via a DOI and with a citation file to encourage people to cite it. What else might we want to do in order to improve how findable, accessible or reusable it is? One further step we could take is to publish the code with a peer reviewed journal. Some traditional journals will accept software submissions, although these are usually as a supplementary material for a paper. There also journals which specialise in research software such as the Journal of Open Research Software, The Journal of Open Source Software or SoftwareX. With these venues, the submission will be the software itself and not a paper, although a short abstract or description of the software is often required.
Working with collaborators
The strength of online collaboration platforms such as GitHub does not just lie in the ability to share code. They also allow us to track problems with that code, for multiple developers to work on it independently and bring their changes together and to review those changes before they are accepted.
Tracking issues with code
A key feature of GitHub (as opposed to Git itself) is the issue tracker. This provides us with a place to keep track of any problems or bugs in the code and to discuss them with other developers. Sometimes advanced users will also use issue trackers of public projects to report problems they are having (and sometimes this is misused by users seeking help using documented features of the program).
The code from the testing chapter earlier has a bug with an extra bracket in eva_data_analysis.py (and if you have fixed that a missing import of summarise_categorical in the test). Let’s go ahead and create a new issue in our GitHub repository to describe this problem. We can find the issue tracker on the “Issues” tab in the top left of the GitHub page for the repository. Click on this and then click on the green “New Issue” button on the right hand side of the screen. We can then enter a title and description of our issue.
A good issue description should include:
- What the problem is, including any error messages that are displayed.
- What version of the software it occurred with.
- Any relevant information about the system running it, for example the operating system being used.
- Versions of any dependent libraries.
- How to reproduce it.
After the issue is created it will be assigned a sequential ID number.
Write an issue to describe our bug
Create a new issue in your repository’s issue tracker by doing the following:
- Go to the GitHub webpage for your code
- Click on the Issues tab
- Click on the “New issue” button
- Enter a title and description for the issue
- Click the “Submit Issue” button to create the issue.
Discussing an issue
Once the issue is created, further discussion can take place with additional comments. These can include code snippets and file attachments such as screenshots or logfiles. We can also reference other issues by writing a # symbol and the number of the other issue. This is sometimes used to identify related issues or if an issue is a duplicate.
Closing an issue
Once an issue is solved then it can be closed. This can be done either by pressing the “Close” button in the GitHub web interface or by making a commit which includes the word “fixes”, “fixed”, “close”, “closed” or “closes” followed by a # symbol and the issue number.
Working in parallel with Git branches
Branching is a feature of Git that allows two or more parallel
streams of work. Commits can be made to one branch without interfering
with another. Branches are commonly used as a way for one developer to
work on a new feature or a bug fix while other developers work on other
features. When those new features or bug fixes are complete, the branch
will be merged back with the main (sometimes called the
master) branch.
Creating a new branch
New Git branches are created with the git branch
command. This should be followed by the name of the branch to create. It
is common practice when the bug we are fixing has a corresponding issue
to name the branch after the issue number and name. For example, we
might call the branch 01-extra-bracket-bug instead of
something less descriptive like bugfix.
For example, the command:
will create a new branch called 01-extra-bracket-bug. We
can view the names of all the branches by running
git branch with no parameters. By default there should be
one branch called main (formerly master) and
our new 01-extra-bracket-bug branch. The command will put
* next to the currently active branch.
OUTPUT
01-extra-bracket-bug
* mainWe can see that creating a new branch has not activated that branch.
To switch branches we can either use the git switch or
git checkout command followed by the branch name. For
example:
To create a branch and change to it in a single command we can use
git switch with the -c option (or
git checkout with the -b option). Note that
git switch command is only available in more recent
versions of Git.
Committing to a branch
Once we have switched to a branch any further commits that are made
will go to that branch. When we run a git commit command we
will see the name of the branch we are committing to in the output of
git commit. Let’s edit our code and fix the lack of default
values bug that we entered into the issue tracker earlier on.
Change your code from:
to:
and now commit it.
In the output of git commit -m the first part of the
output line will show the name of the branch we just made the commit
to.
OUTPUT
[01-extra-brakcet-bug 330a2b1] fixes missing values bug, closes #01 If we now switch back to the main branch our new commit
will no longer be there in the source file or the output of
git log.
And if we go back to the 01-extra-bracket-bug branch it
will re-appear.
If we want to push our changes to a remote such as GitHub we have to
tell the git push command which branch to push to. If the
branch doesn’t exist on the remote (as it currently won’t) then it will
be created.
If we now refresh the GitHub webpage for this repository we should see the bugfix branch has appeared in the list of branches.
If we needed to pull changes from a branch on a remote (for example
if we have made changes on another computer or via GitHub’s web based
editor), then we can specify a branch on a git pull
command.
Merging branches
When we have completed working on a branch (for example fixing a bug)
then we can merge our branch back into the main one (or any other
branch). This is done with the git merge command.
This must be run on the TARGET branch of the merge, so we
will have to use a git switch command to set this.
Now we are back on the main branch we can go ahead and merge the changes from the bugfix branch:
Pull requests
On larger projects we might need to have a code review process before changes are merged, especially before they are merged onto the main branch that might be what is being released as the public version of the software. GitHub has a process for this that is called a pull request. Other Git services such as GitLab have different names for this; GitLab calls them merge requests. Pull requests are situations where one developer requests that another merges code from a branch (or “pull” it from another copy of the repository). The person receiving the request then has the chance to review the code, write comments suggesting changes or even change the code themselves before merging it. It is also very common for automated checks of code to be run on a pull request to ensure the code is of good quality and is passing automated tests.
As a simple example of a pull request, we can now create a pull
request for the changes we made on our 01-extra-bracket-bug
branch and pushed to GitHub earlier on. The GitHub webpage for our
repository will now be saying something like “bugfix had recent pushes
n minutes ago - Compare & Pull request”. Click on this
button and create a new pull request.
Give the pull request a title and write a brief description of it, then click the green “Create pull request” button. GitHub will then check if we can merge this pull request without any problems. We will look at what to do when this is not possible later on.
There should be a green “Merge pull request” button, but if we click on the down arrow inside this button there are three options on how to handle this request:
- Create a merge commit
- Squash and merge
- Rebase and merge
The default is option 1, which will keep all of the commits made on our branch intact. This can be useful for seeing the whole history of our work, but if we’ve done a lot of minor edits or attempts at fixing a problem to fix one bug it can be excessive to have all of this history saved. This is where the second option comes in, this will place all of our changes from the branch into just a single commit, this might be much more obvious to other developers who will now see our bugfix as a single commit in the history. The third option merges the branch histories together in a different way that doesn’t make merges as obvious, this can make the history easier to read but effectively rewrites the commit history and will change the commit hash IDs. Some projects that you contribute to might have their own rules about what kind of merge they will prefer. For the purposes of this exercise we’ll stick with the default merge commit.
Go ahead and click on “Merge pull request”, then “Confirm merge”. The changes will now be merged together. GitHub gives us the option to delete the branch we were working on, since its history is preserved in the main branch there is no reason to keep it.
Using forks instead of branches
A fork is similar to a branch, but instead of it being part of the same repository it is a entirely new copy of the repository. Forks are commonly used by Github users who wish to work on a project that they are not a member of. Typically forking will copy the repository to our own namespace (e.g. github.com/username/reponame instead of github.com/projectname/reponame)
To create a fork on github use the “Fork” button to the right of the repository name. After we create our fork we can make some changes and these could even be on the main branch inside our forked repository. GitHub will track that a fork has been made displays a “Contribute” button to create a pull request back to the original repository. Using this we can request that the changes on our fork are incorporated by the upstream project.
Practice pull requests
Q: Work in pairs for this exercise. Share the GitHub link of your repository with your partner. If you have set your repository to private, you will need to add them as a collaborator. Go to the settings page on your GitHub repository’s webpage, click on Collaborators from the left hand menu and then click the green “Add People” button and enter the GitHub username or email address of your partner. They will get an email and an alert within GitHub to accept your invitation to work on this repository, without doing this they won’t be able to access it.
- Now make a fork of your partners repository.
- Edit the
CITATION.cfffile and add your name to it. - Commit these changes to your fork
- Create a pull request back to the original repository
- Your partner will now receive your pull request and can review
Do not forget to commit any uncommitted changes you may have and then push your work to GitHub.
Further reading
We recommend the following resources for some additional reading on the topic of this episode:
- Licencing and citation episodes from the Software Carpentry’s Git Novice lesson
- Carpentries GitHub Skill ups for instructors and maintainers
- RSG Southampton Git lesson - collaboration section
- Open source definition, by the Open Source Initiative
- What is free software?
Also check the full reference set for the course.
Key Points
- Zenodo can be used to archive a GitHub repository and obtain a DOI for it.
- We include a CITATION file with our code to tell people how to cite it.
- GitHub can help us track bugs or issues with software.
- Git branches can be used to allow multiple developers to work on the same part of code in parallel.
- The
git branchcommand shows the list of branches and can create new branches. - The
git switchcommand changes which branch we are working on. - The
git mergecommand merges another branch into the current one. - Pull requests allow developers to work on their own branch/fork and then request other developers review their changes before they are merged.
Content from Wrap-up
Last updated on 2025-06-17 | Edit this page
Overview
Questions
- What are the FAIR research principles?
- How do FAIR principles apply to software?
- What are the wider Research Software Development Principles and where does FAIR fit into them?
Objectives
After completing this episode, participants should be able to:
- Explain the FAIR research principles in the context of research software
- Explain why these principles are of value in the research community
- Reflect on the Research Software Development Principles and their relevance to own research.
The good development practices taught in this lesson will help you build better research software. Some may require time and perseverance to implement and embed in your routine. Others are small changes you can start practicing today.

FAIR Research Software
One framework that can help you evaluate the alignment of a piece of research software with best practices in reproducibility are the FAIR Principles for Research Software. The practices taught here fall within this framework and it can be a good place to find out what else you can start doing to improve even further.
What is FAIR?
FAIR stands for Findable, Accessible, Interoperable, and Reusable and comprises a set of principles designed to increase the visibility and usefulness of your research to others. The FAIR data principles, first published in 2016, are widely known and applied today. Similar FAIR principles for software have now been defined too. In general, they mean:
- Findable - software and its associated metadata must be easy to discover by humans and machines.
- Accessible - in order to reuse software, the software and its metadata must be retrievable by standard protocols, free and legally usable.
- Interoperable - when interacting with other software it must be done by exchanging data and/or metadata through standardised protocols and application programming interfaces (APIs).
- Reusable - software should be usable (can be executed) and reusable (can be understood, modified, built upon, or incorporated into other software).
Each of the above principles can be achieved by a number of practices listed below. This is not an exact science, and by all means the list below is not exhaustive, but any of the practices that you employ in your research software workflow will bring you closer to the gold standard of fully reproducible research.
Findable
- Create a description of your software to make it discoverable by search engines and other search tools
- Use standards (such as CodeMeta) to describe interoperable metadata for your software (see Research Software Metadata Guidelines)
- Place your software in a public software repository (and ideally register it in a general-purpose or domain-specific software registry)
- Use a unique and persistent identifier (DOI) for your software (e.g. by depositing your code on Zenodo), which is also useful for citations - note that depositing your data/code on GitHub and similar software repositories may not be enough as they may change their open access model or disappear completely in the future, so archiving your code means it stands a better chance at being preserved
Accessible
- Make sure people can obtain get a copy your software using standard communication protocols (e.g. HTTP(S), (S)FTP, etc.)
- The code and its description (metadata) has to be available even when the software is no longer actively developed (this includes earlier versions of the software)
Interoperable
- Use community-agreed standard formats for inputs and outputs of your software and its metadata
- Communicate with other software and tools via standard protocols and APIs
Reusable
- Document your software (including its functionality, how to install and run it) so it is both usable (can be executed) and reusable (can be understood, modified, built upon, or incorporated into other software)
- Give a licence to your software clearly stating how it can be reused
FAIR is a process, not a perfect metric
FAIR is not a binary metric - there is no such thing as “FAIR or”not FAIR”.
FAIR is not a perfect metric, nor does it provide a full and exhaustive software quality checklist - there are other good software quality practices not covered by FAIR. Conversely, software may be FAIR but still not very good in terms of its functionality.
FAIR is not meant to criticise or discredit work.
FAIR refers to the specific values of and describes a set of principles to aid open and reproducible research that can be a helpful guide for researchers who want to improve their practices (by helping them see where they are on the FAIR spectrum and help them on a journey to make their software more FAIR).
Assessing the FAIRness of your Research Software
Here are some questions to help you assess where on the FAIR spectrum your software is.
- Findable
- If these files were emailed to you, or sent on a chat platform, or handed to you on a memory stick, how easy would it be to find them again in 6 months, or 3 years?
- If you asked your collaborator to give you the files again later on, how would you describe them? Do they have a clear name?
- If more data was added to the data set later, could you explain exactly which data you used in the original analysis?
- Accessible
- If the person who gave you the files left your institution, how would you get access to the files again?
- Once you have the files, can you understand the code? Does it make sense to you?
- Do you need to log into anything to use this? Does it require purchase or subscription to a service, platform or tool?
- Interoperable
- Is it clear what kind of input data it can read and what kind of output data is produced? Will you be able to create the input files and read the output files with the tools your community generally uses?
- If you wanted to use this tool as part of a larger data processing pipeline, does it allow you to link it with other tools in standard ways such as an API or command-line interface?
- Reusable
- Can you run the code on your platform/operating system (is there documentation that covers installation instructions)? What programs or libraries do you need to install to make it work (and which versions)? Are these commonly used tools in your field?
- Do you have explicit permission to use your collaborators code in your own research and do they expect credit of some form (paper authorship, citation or acknowledgement)? Are you allowed to edit, publish or share the files with others?
- Is the language used familiar to you and people in your research field? Can you read the variable names in the code and the column names in the data file and understand what they mean?
- Is the code written in a way that allows you to easily modify or extend it? Can you easily see what parameters to change to make it calculate a different statistic, or run on a different input file?
FAIR Software is Better Software
You may or may not find the FAIR Research Software Principles a helpful way of framing good practices. The important thing is to focus on how adopting these individual ways of working contributes to making your software better. Many of the practices taught here – and advocated for in the FAIR framework – will make your improve your life as a researcher and software developer.
The table below provides a summary of some good practices for developing research software, together with different tools that can help with such practices and how they contribute to the the FAIR and other good software principles.
| Practices | Tools | FAIR | Readable/Understandable | Correct/Reliable | Sustainable/Maintainable |
|---|---|---|---|---|---|
| Use version control |
git, GitHub, GitLab, BitBucket |
F | |||
| Write modular code with well defined interfaces | R | x | x | ||
| Connect reusable software components into automated/reproducible software pipelines | Command line tools, CLI, workflow tools (Galaxy, Snakemake, WorkflowHub, CWL) | I, R | |||
| Use reproducible software development environments |
venv, conda, IDEs (integration with
virtual envs.), Docker, etc. |
R | |||
| Use conventional folder structures, format your code to comply with coding conventions | PEP8, IDEs (help with formatting and conforming with coding conventions) | R | |||
| Use standard exchange data formats/communication protocols/interfaces | CSV, YAML, JSON, CLI, REST, HTTP(S), etc. | I | x | ||
| Test your software | Unit, functional, integration, regression, etc. tests, IDEs (testing and debugging), CI/CD (automation) | x | |||
| Document your software | Comments and documentation strings, README, guides, contributions guidelines | R | x | ||
| Share your software & encourage review | Code sharing platforms/services (e.g. GitHub, GitLab, BitBucket) and their code review facilities | F, A | x | ||
| License your software | Various open source licences for code (MIT, BSD, GPL, LGPL, etc.), LICENSE | R | |||
| Use persistent identifiers for your software | DOIs, SWHIDs, Zenodo, FigShare, Software Heritage, etc. | F | x | ||
| Provide citation & metadata info for your software | CITATION, DOIs, Zenodo, CFF, cffinit, CodeMeta,
etc. |
R | x | ||
| Build community & encourage collaboration around your software | Code of Conduct, README, CONTRIBUTING, open source project governance processes | x |
Best practices are always evolving and there is usually more we could be doing to make our software even better, even more reproducible, even FAIRer. The skills and techniques introduced in this lesson are a great place to start!
Tools for assessing FAIRness
Here are some tools that can check your software and provide an assessment of its FAIRness:
Research software development principles
Software and people who develop it have significance within the research environment and a broader impact on society and the planet. FAIR research software principles cover some aspects and operate within the wider Research Software Development Principles - recommended by Software Sustainability Institute’s Director Neil Chue Hong during his keynote at RSECon23. These principles can help us explore and reflect on our own work and guide us on a path to better research.
Further reading
We recommend the following resources for some additional reading on FAIR Research Software:
- “Five recommendations for FAIR software”
- “10 easy things to make your research software FAIR”
- “Ten simple rules for training scientists to make better software”
- Automating assessment of the FAIR Principles for Research Software (FAIR4RS)
- Short online courses on various aspects of research software (including FAIR research software and data), by the NeSC Research Software Support
- CodeRefinery - training and e-Infrastructure for research software development
- A self-assessment checklist for FAIR research software, by the Netherlands eScience Center and Australian Research Data Commons
- Awesome Research Software Registries - a list of research software registries (by country, organisation, domain and programming language) where research software can be registered to help promote its discovery
Please check out the following resources for some additional reading on the topic of this course and the full reference set.
Key Points
- FAIR data and software is Findable, Accessible, Interoperable, Reusable.
- These principles support research and researchers by saving time, reducing barriers to discovery, and increasing impact of the research output.
- When developing software for your research, think about how it will help you and your team, your peers and domain/community and the world.



