AI for Automation
Overview
Teaching: 10 min
Exercises: 2 minQuestions
How is AI used for automating tasks in biomedical experimental setups?
What are examples of biomedical AI-driven software packages and what can they be used for?
Objectives
Learning about biomedical examples of AI automation
Developing case studies for your research
Outline:
In the previous episode, we discussed the history of AI and the three broad categories – simulation, symbolic AI, and machine learning. When it comes to applications of machine learning and AI in biomedical and life sciences, we can divide those by application into two main categories: automation and insight.
In this episode, we will look at the following:
- AI applications to automate analytical processes
- Case studies/ Examples: Entrez, QUPATH, MicrobeJ, Cellpose, DeepLabCut
Using Automation in Research:
For automation, AI is used to replace tasks that would take a researcher a long time to do manually, in order to speed up workflow outputs. This can be invaluable during data processing, freeing up a researcher’s time for other tasks and possibly removing human error.
Automating Tasks
Automation is ubiquitous to our everyday experiences in working with computers–from simply typing words to creating figures, or spellcheck and autocorrect while messaging. Using AI can optimise tasks that are necessary for research or experimentation.
For example, the field of computer vision is about automating the task of counting and/or distinguishing visual elements in images or photographs. Computer vision methods are being applied in biomedical image data, for counting cells on a slide or tracking a mouse’s locomotion on video.
Many open-source tools have been produced and are already in use across the biomedical sciences. In this episode, we will introduce a range of open-source AI tools. Contrary to proprietary software, which is often rigid and opaque, open-source tools are developed in close collaboration with and contributions by their user community.
Not being at the cutting edge of AI development doesn’t mean we can’t also benefit from the AI tools that can assist with research tasks.
Case Study: The Molecular Biology Database Entrez
Website: Last edit from 2004: https://www.ncbi.nlm.nih.gov/Web/Search/entrezfs.html redirect from https://www.ncbi.nlm.nih.gov/Entrez/ to https://www.ncbi.nlm.nih.gov/search/ // https://www.tutorialspoint.com/biopython/biopython_entrez_database.htm

The ability to search and look up molecular biology databases is an example of automation.
Entrez is an online search system provided by NCBI. It provides access to nearly all known molecular biology databases with an integrated global query supporting Boolean operators and field search. It returns results from all the databases with information like the number of hits from each database, records with links to the originating database, etc.
Some of the popular databases which can be accessed through Entrez are listed below −
- Pubmed
- Pubmed Central
- Nucleotide (GenBank Sequence Database)
- Protein (Sequence Database)
- Genome (Whole Genome Database)
- Structure (Three Dimensional Macromolecular Structure)
- Taxonomy (Organisms in GenBank)
- SNP (Single Nucleotide Polymorphism)
- UniGene (Gene Oriented Clusters of Transcript Sequences)
- CDD (Conserved Protein Domain Database)
- 3D Domains (Domains from Entrez Structure)
Case Study: QUPATH
Website: https://qupath.github.io/ // documentation: https://qupath.readthedocs.io/en/0.4/ //
Reference: Bankhead, P., Loughrey, M.B., Fernández, J.A. et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878 (2017). https://doi.org/10.1038/s41598-017-17204-5
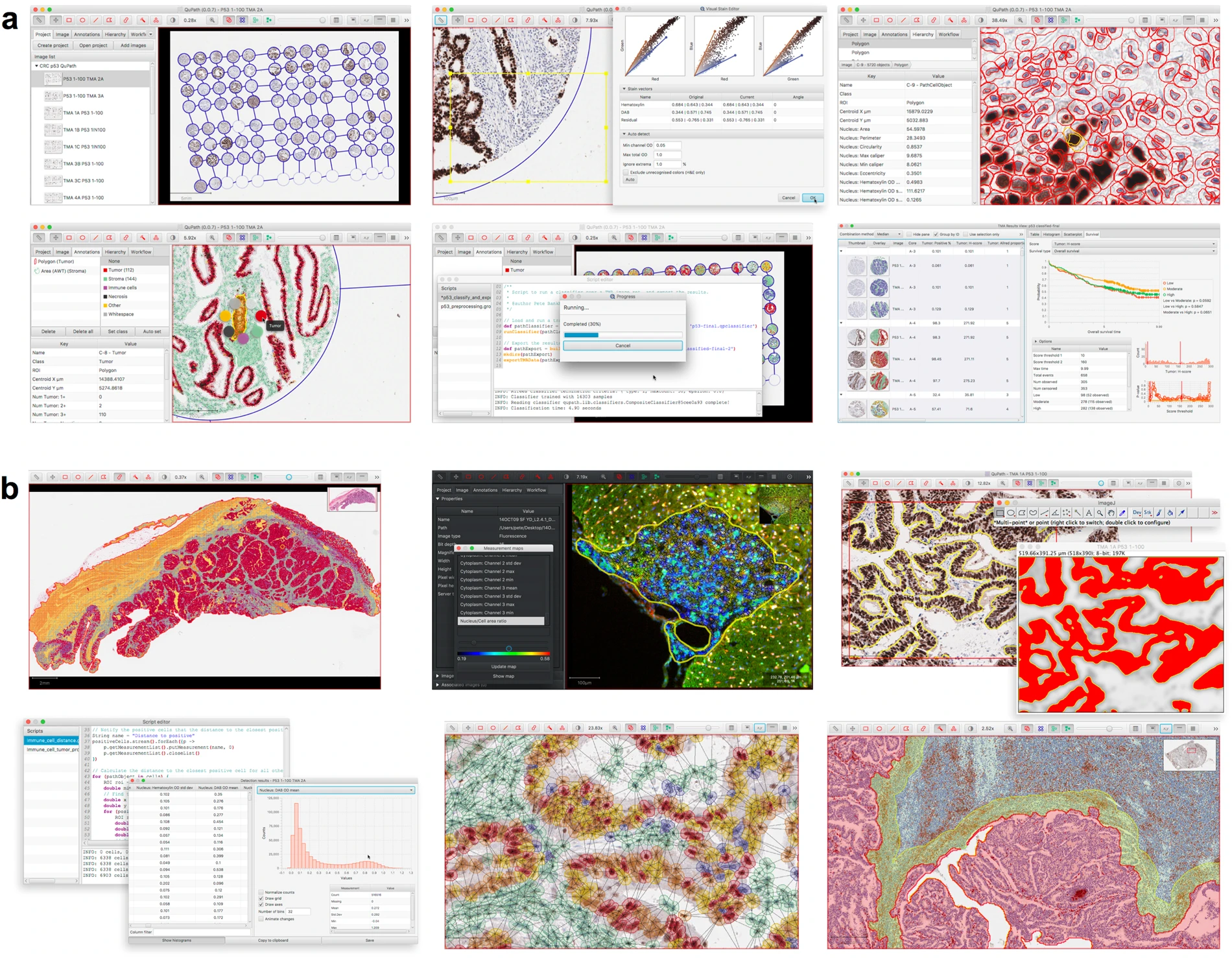
Qupath is a tool using machine learning to work with image analysis in the growing field of “digital pathology”. Scanners can quickly produce tens of gigabytes of image data of tissue slides, and traditional manual assessment is no longer suitable. As a result, automation through software provides a much faster solution.
In recent years, a vibrant ecosystem of open source bioimage analysis software has developed. Led by ImageJ, researchers in multiple disciplines can now choose from a selection of powerful tools, such as Fiji, Icy, and CellProfiler, to perform their image analyses. These open source packages encourage users to engage in further development and sharing of customized analysis solutions in the form of plugins, scripts, pipelines or workflows – enhancing the quality and reproducibility of research, particularly in the fields of microscopy and high-content imaging. This template for open-source development of software has provided opportunities for image analysis to add considerably to translational research by enabling the development of the bespoke analytical methods required to address specific and emerging needs, which are often beyond the scope of existing commercial applications.
Bankhead, P., Loughrey, M.B., Fernández, J.A. et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878 (2017). https://doi.org/10.1038/s41598-017-17204-5
Case Study: MicrobeJ
Website: https://www.microbej.com/
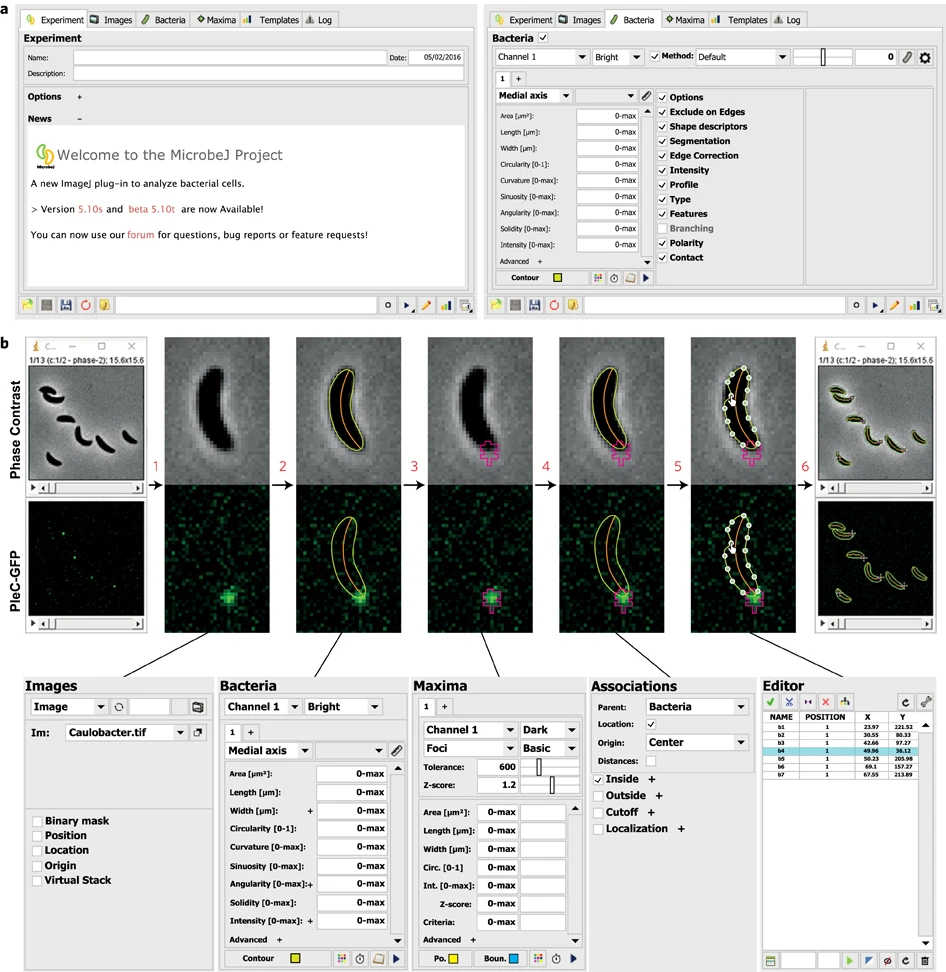
MicrobeJ is a plug-in for the open-source platform ImageJ. MicrobeJ provides a comprehensive framework to process images derived from a wide variety of microscopy experiments with special emphasis on large image sets. It performs the most common intensity and morphology measurements as well as customized detection of poles, septa, fluorescent foci and organelles, determines their subcellular localization with subpixel resolution, and tracks them over time.
Ducret, A., Quardokus, E. & Brun, Y. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1, 16077 (2016). https://doi.org/10.1038/nmicrobiol.2016.77
Case Study: Cellpose
Website: https://www.cellpose.org/
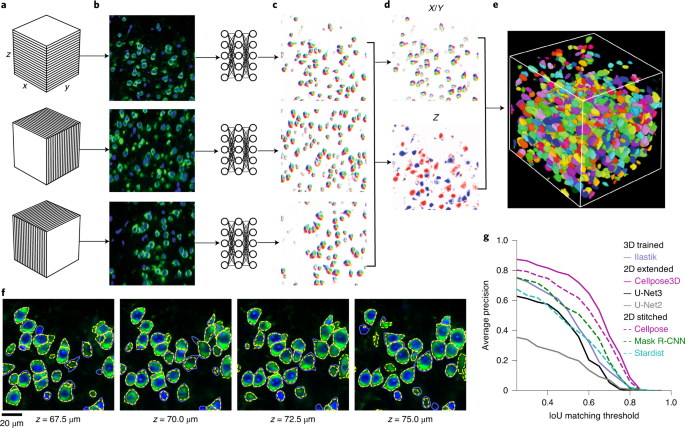
A generalist, deep learning-based segmentation method called Cellpose, which can precisely segment cells from a wide range of image types and does not require model retraining or parameter adjustments. Cellpose was trained on a new dataset of highly varied images of cells, containing over 70,000 segmented objects. We also demonstrate a three-dimensional (3D) extension of Cellpose that reuses the two-dimensional (2D) model and does not require 3D-labeled data. To support community contributions to the training data, we developed software for manual labelling and for the curation of automated results. Periodically retraining the model on the community-contributed data will ensure that Cellpose improves constantly.
Stringer, C., Wang, T., Michaelos, M. et al. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18, 100–106 (2021). https://doi.org/10.1038/s41592-020-01018-x
Case Study: DeepLabCut
Website: http://www.mackenziemathislab.org/deeplabcut // https://github.com/DeepLabCut/DeepLabCut // https://deeplabcut.github.io/DeepLabCut/docs/standardDeepLabCut_UserGuide.html
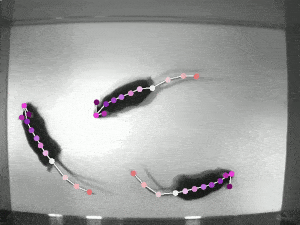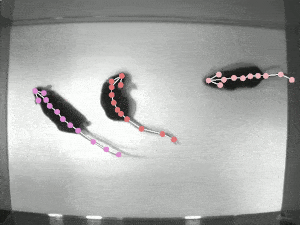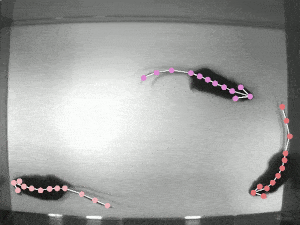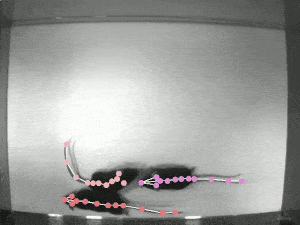
Quantifying behaviour is crucial for many applications in neuroscience, ethology, genetics, medicine, and biology. Videography provides easy methods for the observation and recording of animal behaviour in diverse settings, yet extracting particular aspects of a behaviour for further analysis can be highly time-consuming.
DeepLabCut offers an efficient method for 3D markerless pose estimation based on transfer learning with deep neural networks that achieves excellent results (i.e. you can match human labelling accuracy) with minimal training data (typically 50-200 frames). We demonstrate the versatility of this framework by tracking various body parts in multiple species across a broad collection of behaviours. The package is open source, fast, robust, and can be used to compute 3D pose estimates.
Nath, T., Mathis, A., Chen, A.C. et al. Using DeepLabCut for 3D markerless pose estimation across species and behaviours. Nat Protoc 14, 2152–2176 (2019). https://doi.org/10.1038/s41596-019-0176-0
Exercises
- What tasks can be automated (and how) in your research workflow?
- Which of the presented use cases could be applied in your own experimental setup? If not, why?
Key Points
Despite not being at the cutting edge of AI development we can still benefit from elevated efficiency and accuracy of research data processing.
Examples of AI applications for automation include database search, image analysis and motion tracking
While not every researcher works with generating AI algorithms and models, there are plenty of tools with numerous applications in research that use machine learning. Automating tasks results in larger data sets and less manual work, and it is worth joining the online communities that work on the tools showcased here.