Summary and Setup
Welcome to the Structural Neuroimaging Analysis in Python workshop!
The primary goals of this workshop are to:
- Understand the basics of structural MR image acquisition
- Familiarize yourself with structural MR image (pre)processing pipelines
- Perform and visualize group-level neuroanatomical analyses
Things to keep in mind:
Magnetic resonance (MR) imaging is a medical imaging technique used to visualize anatomy and the physiological processes of the body. MR imaging scanners use strong magnetic fields, magnetic field gradients, and radio waves to generate images of the organs in the body.
In structural neuroimaging, MR scans can refer to several different image modalities including, T1-weighted, T2-weighted, diffusion weighted images (DWI), Proton-Density (PD), Fluid attenuation inversion recovery (FLAIR) etc.
An MR (pre)processing pipeline is a set of sequential image processing tasks performed on acquired MR scans prior to the statistical analysis.
MR software packages: In order to standardize and simplify computational effort, several software packages encapsulate MR (pre)processing pipelines. Thus as a user, you need not know the details of each image processing algorithm. Nevertheless it is useful to understand the key objectives of these tasks, the corresponding computational approaches, and their impact on the downstream analyses. This will 1) help developers to improve the underlying algorithms and 2) help users to customize the neuroimaging pipelines according to specific dataset requirements. Here are a few common software packages:
- FreeSurfer
- FSL
- CIVET
- ANTs
- SPM (mostly for functional data)
- AFNI (mostly for functional data)
- sMRIprep or fMRIprep (pipeline customization)
Note: All of this may sound complicated, but we’ll explain things step-by-step in depth with practical examples as the course goes along. We will begin our computational journey starting from how an MR image is acquired, followed by several pre-processing tasks, with the end goal of conducting a statistical analysis to investigate neuroanatomical differences between patients and healthy control groups.
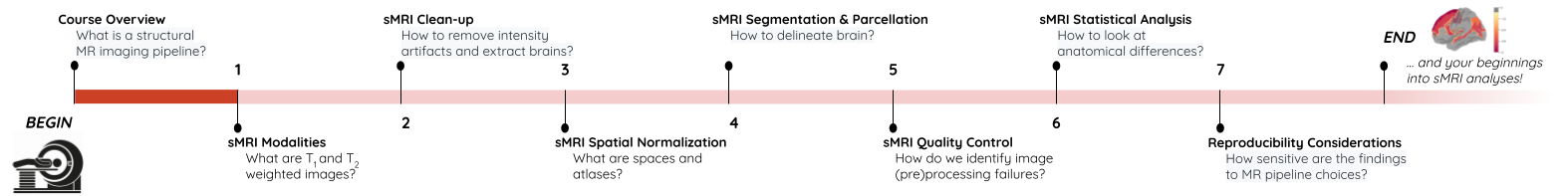
You Are Here!
Prerequisites
Attendees must have some base familiarity with Python and NIfTI images in order to comfortably progress through the lesson
Setting up the tutorial environment
Getting workshop material
Method 1: Downloading directly from the repository
On the GitHub repo, click the green button that says “Code”, then click Download ZIP. Once downloaded, extract the ZIP file.
Method 2: Using Git
Using this method requires a (very) useful piece of software called
git. The process of installing git depends heavily on
whether you’re using MacOS, Windows or Linux. Follow the instructions in
the link below to set up git on your PC:
Once you’ve installed git, open up your terminal and do
the following:
git clone https://github.com/carpentries-incubator/SDC-BIDS-sMRI.gitThis will download the repository directly into your current directory.
Setting up Python environment
We use python version 3.6.0, but any newer version should also work (Python 2 versions haven’t been tested). There are many methods to setting up a python environment but we’d recommend using some sort of virtual environment as to not break your system python install. Two methods (of many) are listed below:
Method 1: Setting up conda environment (easiest) [Windows, Linux, MacOS]
For easy set-up we recommend Anaconda to manage python packages for scientific computing. Once installed, setting up the python environment can be done quite easily:
Windows
- Install Anaconda Python version 3.7
- Open Anaconda Navigator
- Click on Environments on the left pane
- Click Create then type in
SDC-BIDS-sMRI - In the
SDC-BIDS-sMRIentry click the play button then click Open Terminal - In terminal type:
conda install -y numpy pandas scipy scikit-learn matplotlib seaborn jupyter ipykernel nb_conda
conda install -y -c conda-forge awscli lxml nilearn nibabel statsmodels
pip install pybids- Close the terminal, click on the play button again and open Jupyter Notebook
- Navigate to
SDC-BIDS-sMRIfolder you downloaded earlier. - Done!
Linux and MacOS
After installing Anaconda, open terminal and type:
cd SDC-BIDS-sMRI
conda create -p ./SDC_sMRI_workshop_2021
source activate $(pwd)/SDC_sMRI_workshop_2021
conda install numpy pandas scipy scikit-learn matplotlib seaborn jupyter ipykernel nb_conda
conda install -c conda-forge awscli nibabel nilearn lxml statsmodels
pip install pybidsMethod 2: Using pyenv [Linux, MacOS]
An alternative method uses pyenv with pyenv virtualenv. This is a favourite because it seamlessly integrates multiple python versions and environments into your system while maintaining use of pip (instead of conda).
cd SDC-BIDS-sMRI
pyenv virtualenv 3.6.0 SDC_sMRI_workshop_2021
echo SDC_sMRI_workshop_2021 > .python-version
pip install --requirement requirements.txtFinally open up the jupyter notebook to explore the tutorials:
cd SDC-BIDS-sMRI
# Include the line below if you are using anaconda environment and it is not already active
# source activate $(pwd)/SDC_sMRI_workshop_2021
jupyter notebook
Data used by the notebooks
All datasets needed for the notebook examples are inside the ./SDC-BIDS-sMRI/local_data/ directory.
