From the scanner to our computer
Last updated on 2025-02-26 | Edit this page
Overview
Questions
- What are the main MRI modalities?
- What’s the first step necessary to start working with MRI data?
Objectives
- Understand how different MRI modalities differ and what each one represents
- Become familiar with converting MRI data from DICOM to NIfTI
Types of MR scans
Anatomical
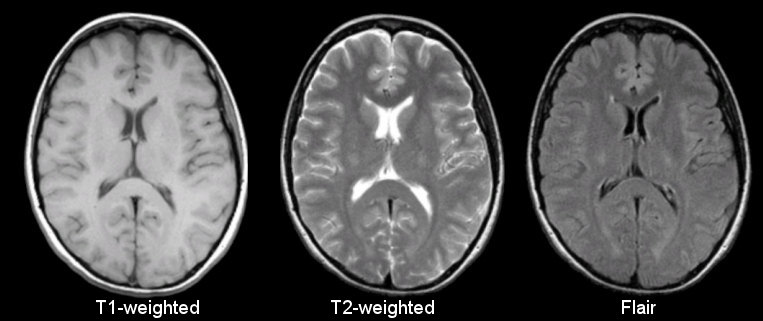
Sourced from https://case.edu/med/neurology/NR/MRI%20Basics.htm
- 3D image of anatomy (i.e., shape, volume, cortical thickness, brain region)
- can differentiate tissue types
Functional
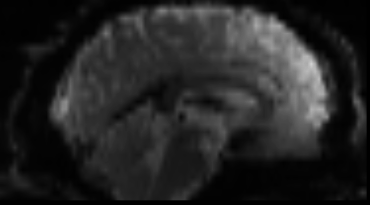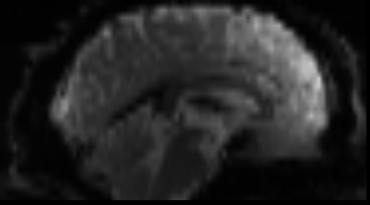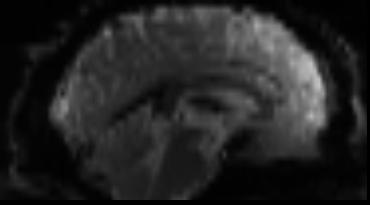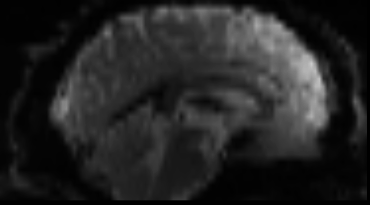
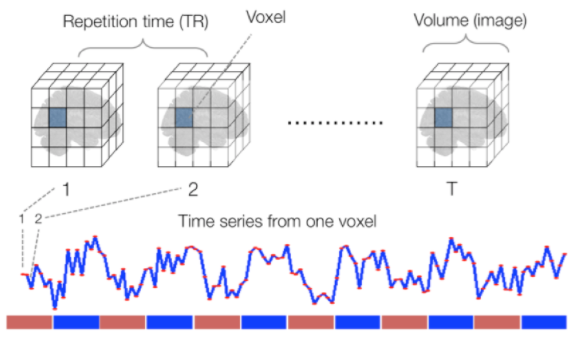
Sourced from Wagner and Lindquist, 2015
- tracks the blood oxygen level-dependant (BOLD) signal as an analogue of brain activity
- 4D image (x, y, z + time)
Diffusion

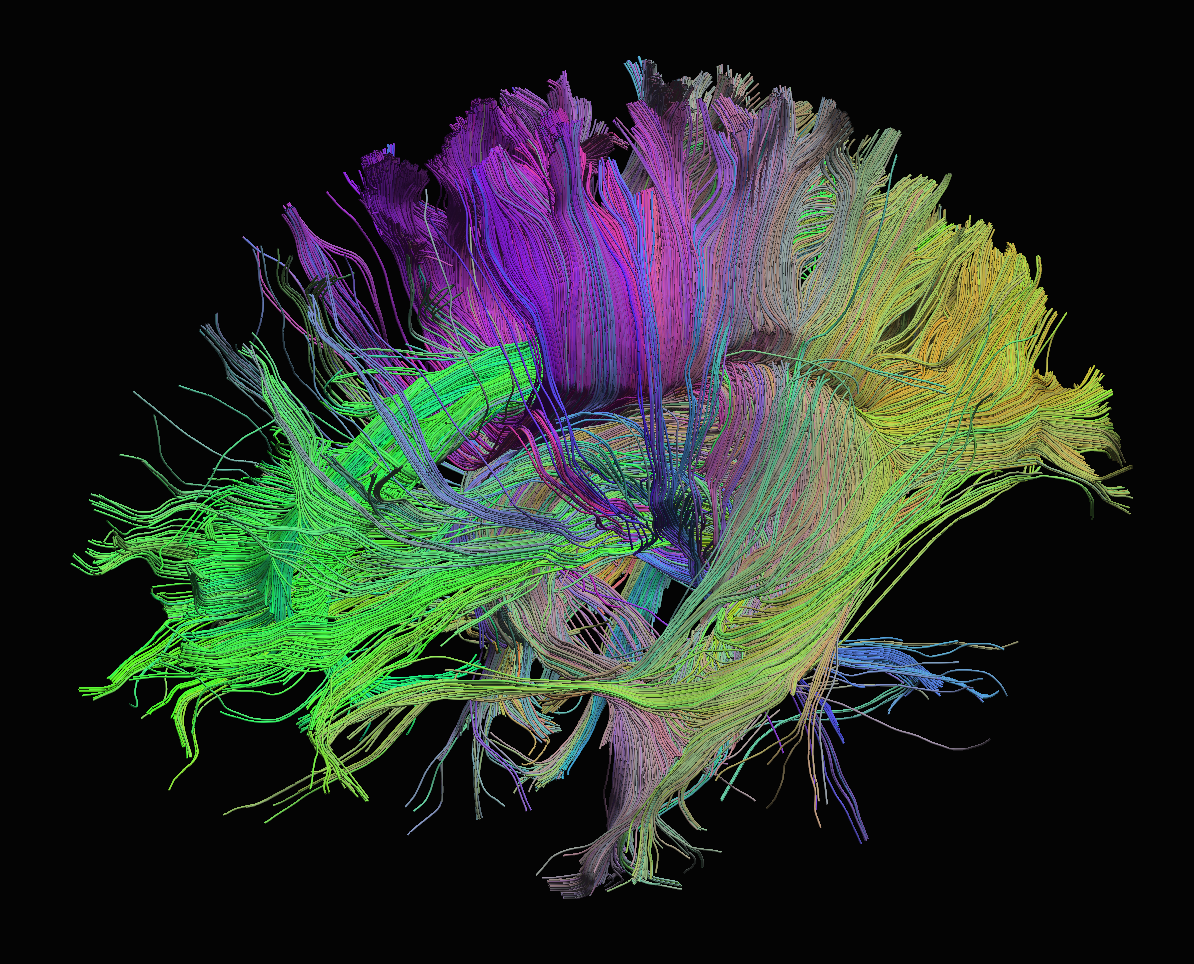
Sourced from http://brainsuite.org/processing/diffusion/tractography/
- measures diffusion of water in order to model tissue microstructure
- 4D image (x, y, z + direction of diffusion)
- need parameters about the strength of the diffusion “gradient” and
its direction (
.bvaland.bvec)
Neuroimaging file formats
Common file formats:
| Format Name | File Extension | Origin/Group |
|---|---|---|
| DICOM | none (*) | ACR/NEMA Consortium |
| Analyze |
.img/.hdr
|
Analyze Software, Mayo Clinic |
| NIfTI |
.nii (**) |
Neuroimaging Informatics Technology Initiative |
| MINC | .mnc |
Montreal Neurological Institute |
| NRRD | .nrrd |
Gordon L. Kindlmann |
| MGH |
.mgz or .mgh
|
Massachusetts General Hospital |
(*) DICOM files typically have a .dcm file extension.
(**) Some files are typically compressed and their extension may
incorporate the corresponding compression tool extension
(e.g. .nii.gz).
From the MRI scanner, images are initially collected in the DICOM format and can be converted to these other formats to make working with the data easier.
Let’s download some example DICOM data to see what it looks like. This data was generously shared publicly by the Princeton Handbook for Reproducible Neuroimaging.
BASH
wget https://zenodo.org/record/3677090/files/0219191_mystudy-0219-1114.tar.gz -O ../data/0219191_mystudy-0219-1114.tar.gz
mkdir -p ../data/dicom_examples
tar -xvzf ../data/0219191_mystudy-0219-1114.tar.gz -C ../data/dicom_examples
gzip -d ../data/dicom_examples/0219191_mystudy-0219-1114/dcm/*dcm.gz
rm ../data/0219191_mystudy-0219-1114.tar.gz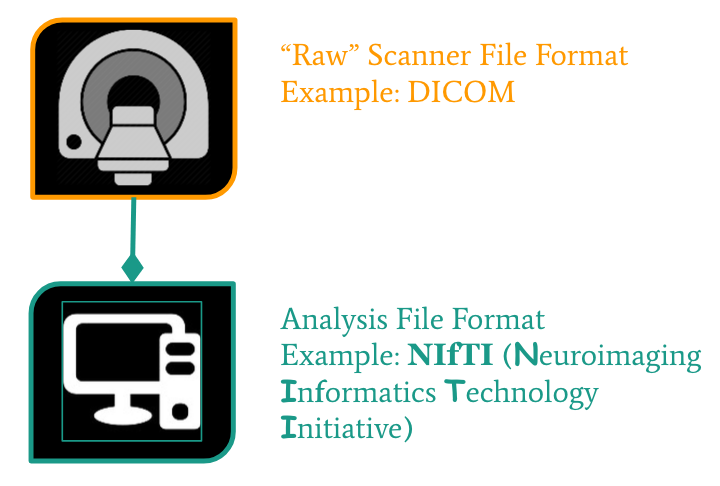
NIfTI is one of the most ubiquitous file formats for storing neuroimaging data. If you’re interested in learning more about NIfTI images, we highly recommend this blog post about the NIfTI format. We can convert our DICOM data to NIfTI using dcm2niix.
We can learn how to run dcm2niix by taking a look at its
help menu.
Converting DICOM to NIfTI
Convert the Princeton DICOM data to NIfTI
- MRI can capture anatomical (structural), functional, or diffusion features.
- A number of file formats exist to store neuroimaging data.
