Introduction: Machine Learning Ready RNA-Seq Data
Last updated on 2024-05-14 | Edit this page
Estimated time: 11 minutes
Overview
Questions
- Where can I find a publicly available RNA-Seq dataset suitable for a machine learning analysis?
- What format is RNA-Seq data stored in on public repositories?
- What characteristics of a dataset do I need to consider to make it ‘ready’ for a machine learning /AI modelling analysis?
Objectives
- Recall the main platforms hosting functional genomic datasets
- Explain the difference between raw and processed RNA-Seq data, and the different stages of processing for RNA-Seq data
- Describe the factors that must be considered in readying an RNA-Seq dataset for a machine learning analysis?
Functional Genomics platforms
There are two major public repositories for sourcing public functional genomics data sets, in particular microarray and next-generation sequencing data. Here we focus on NGS data, in particular RNA-Seq transcriptomics datasets:
The Array Express collection within the Bio Studies database, maintained by the European Molecular Biology Laboratory - European Bioinformatics Institute (EMBL-EBI).
The Gene Expression Onmibus (GEO), maintained by the National Center for Biotechnology Information (NCBI), part of the US National Institues of Health (NIH).
Each database contains thousands of RNA-Seq datasets relating to a wide variety of experiments, not all of which would be suitable for analysis with a machine learning approach.
Raw vs. Processed RNA-Seq Data
Public datasets will contain both raw and processed RNA-Seq data, as well as information about the relationship between the RNA-Seq data and study samples, and the design of the experiment. Here is a brief summary of the difference between raw and processed RNA-Seq data:
| Data Type | What it is | File Format |
|---|---|---|
| Raw data | Sequences of each read obtained from the sequencing instrument, along with quality scores for each sequence | FASTQ |
| Processed data | Matrix of abundance values (e.g. integer counts) for the feature of interest, which may be genes, transcripts, exons or miRNAs | .csv / .txt / .tsv |
For more information on the fastq file format, see RDMBites | FASTQ Format and for information on the processing of raw fastq to gene abundance data, see RDMbites | RNAseq expression data.
Callout
The description “processed” means different things in different studies. Things to be aware of:
- Data is often generated and made available at different levels:
- Raw abundance of sequencing reads for each feature of interest (e.g. integer counts of the number of reads mapped to a given gene)
- Normalised abundance, further processed to account for sequence depth and/or gene length such as TPM and FPKM (output using DESeq2, edgeR)
- Normalised and transformed adundance with normalisation and further transformation applied such as log2 or variance stabilising transformation (vst) applied
The features of interest, say transcripts, may also have been filtered to remove transcripts with read counts below a particular threshold.
Transcript level information may have been mapped to genes, and the expression for multiple transcripts summarised (average, maximum) at the gene level
The data may be given as a matrix of features of interest for all samples, or alternatively as a separate file for each sample.
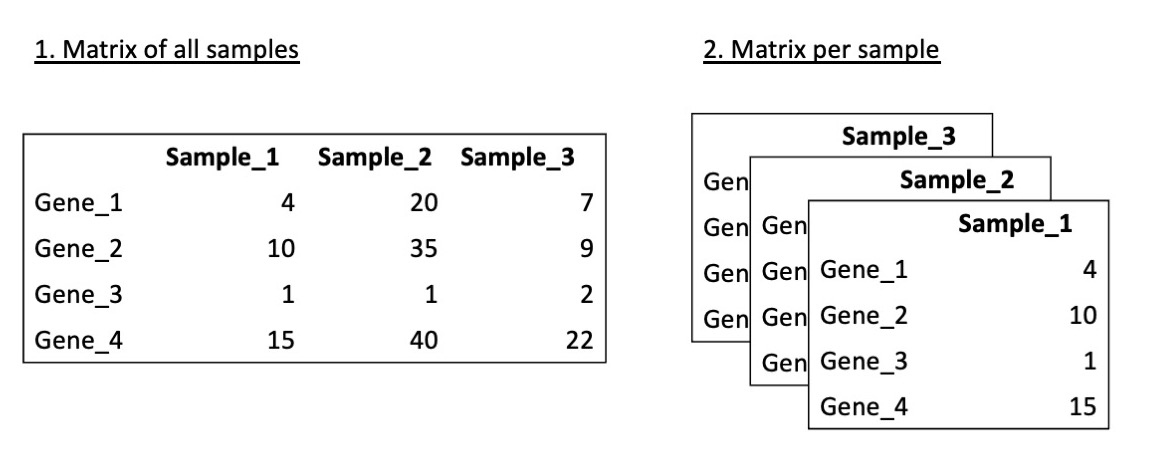
It is important to carefully read the protocols to understand determine how the data has been processed and if is appropriate for your task.
The starting point for a machine learning model is processed RNA-Seq data, in the form of count data. This could be raw count data, or TPM or FPKM pre-processed data.
Dataset Characteristics for supervised learning
Machine learning / AI modelling is a powerful tool in the analysis of RNA-Seq gene expression data. ML techniques are widely used to discover new biomarkers for disease diagnosis and treatment monitoring, and to discover hidden patterns in gene expression that enhance our understanding of the underlying biological pathways. The success of a ML/AI model depends heavily on the input data. Finding an appropriate dataset can be a challenge and the data must be selected carefully as a predictive model trained on the unreliable or inappropriate data will produce unreliable predictions.
There is currently no curated “ML/AI ready” datasets that meet the requirements of machine learning analyses within public functional genomics repositories. It is therefore important to examine a data set carefully to confirm its appropriateness for a supervised machine learning task. The following characteristics of the data should be considered:
| Characteristic | Considerations |
|---|---|
| Quality and Provenance | Assess source of the data, its likely quality and whether it is recognised by the community. Datasets must have sufficient metadata, ideally connected to a domain-specific or community-specific ontology so that the meaning is widely interpretable and comparable between studies. |
| Size | Ensure the data set captures the full complexity of the underlying distribution. If there is significant biological heterogeneity in the sample population, will there be sufficient samples to create independent and representative train, validation and tests sets? Bulk RNA-Seq datasets with only a few samples for each group are unlikely to be sufficient to train a machine learning classifier. |
| Format and Integrity | Machine learning models require data to be machine readable and input in a specific format. Data collected experimentally and data acquired from public repositories will likely need to be reformatted and checked for integrity issues. |
| Technical Noise | RNA-Seq data is likely to contain some technical noise resulting from experimental processes that is unrelated to the underlying biology. This has the potential to bias machine learning algorithms and should be identified and ideally removed. |
| Distribution and Scale | Many machine learning algorithms are sensitive to the distribution and the scale of the input data. RNA-Seq data must be transformed and re-scaled to perform well in a machine learning analysis. |
In the following episodes, we will address each of these issues in turn, working through the collection of a RNA-Seq dataset, reformatting, integrity checking, noise elimination, transformation and rescaling to make the data ready for machine learning. Further information on some of the data considerations in supervised machine learning with biological data can be found in the DOME recommendations.
Key Points
- The two main repositories for sourcing RNA-Seq datasets are ArrayExpress and GEO
- Processed data, in the form of raw counts, or further processed counts data is the starting point for machine learning analysis
- Sourcing and appropriate RNA-Seq dataset and preparing it for a machine learning analysis requires you to consider the dataset quality, size, format, noise content, data distribution and scale
