Workshop Structure
This Workshop is divided in 4 lessons:
- Introduction to the Command Line for Pangenomics.
- Introduction to Python.
- Pangenome Analysis in Prokaryotes.
- Topological Data Analysis for Pangenomics.
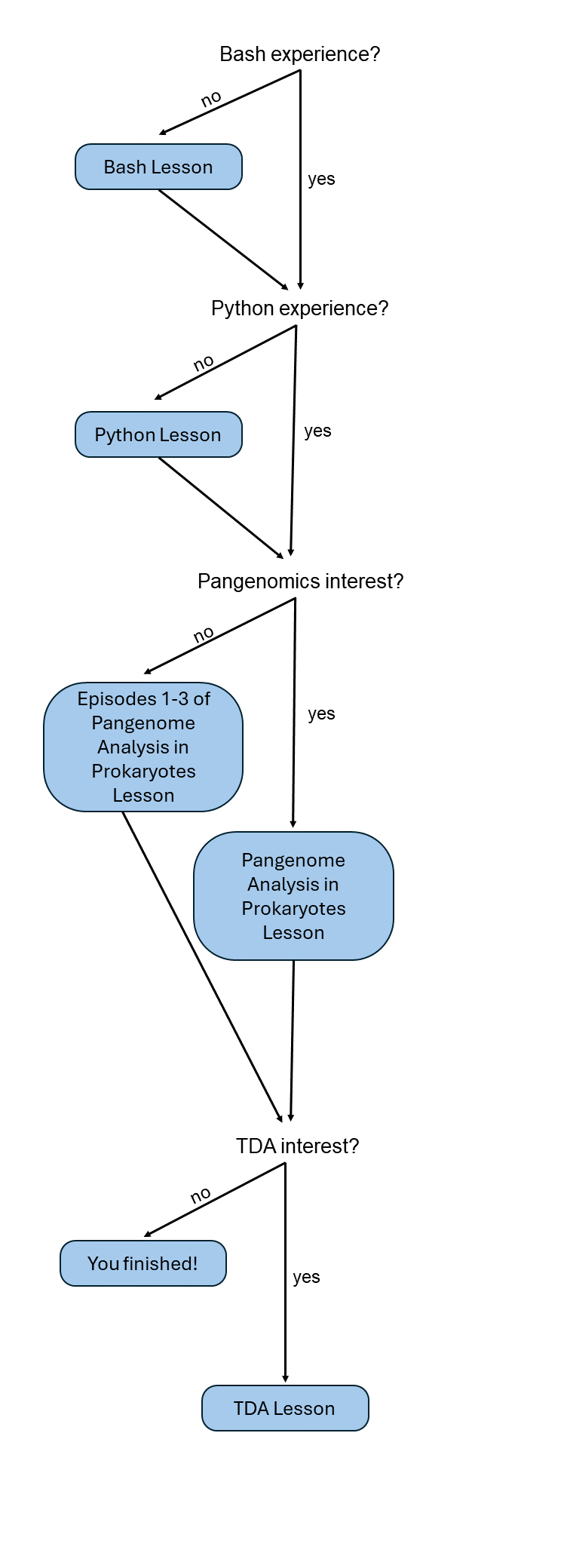
The sixteen-hour workshop is designed for a whole weekend. Nevertheless, it could be a reference manual for students learning Pangenomics or TDA. The workshop is divided in 1) an introduction to Command-Line and Python for those learners with no previous coding experiences and 2) two topic lessons, including pangenome analysis and topological data analysis.
Lessons 1 and 2, which cover an introduction to Bash and Python, are optional. If you already have previous knowledge of these languages, you can start directly with Lesson 3 on pangenome Analysis in Prokaryotes. Lesson 4 introduces topological data analysis and its applications to pangenomics. One short way to follow the workshop in less time, but without losing the main idea, is to complete episodes 1, 2, and 3 of Lesson 3, and then move on to the TDA lesson. With this path, only a fraction of the pangenome is obtained, “the resistome”, i.e., genes annotated as conferring antibiotic resistance. The resistome is a good first example of applying TDA on genomic features.
Episodes 4 (Measuring Sequence Similarity) and 5 (Clustering with BLAST Results) of Lesson Pangenome Analysis in Prokaryotes introduce constructing a pangenome using a test set of genes rather than the complete dataset. This test or mini pangenome is utilized in the chapters of the TDA section. However, in those chapters, you will also find a link to the database of this mini pangenome in case you haven’t completed these episodes. These episodes use Python, so you can refer to the introductory Python lesson if you are unfamiliar with it.
Starting in episode 6, you will find various software tools for constructing a pangenome, each with a different purpose. You can follow just one or all of these chapters to explore different approaches. Episode 7 depends on a file obtained in Episode 6; however, you can skip this file by replacing the first instruction in the Gene Cluster subsection with ppanggolin cluster -p pangenome.h5 --cpu 8. If you replace this instruction, your results will differ from those in the episode, and you will need to interpret them accordingly. Nonetheless, it is possible to skip this step.
In each episode of every lesson, you will find several exercises labeled as Beginner, Intermediate, or Advanced; if you are a beginner in the episode contents, start with the beginner exercise and then move to the Intermediate or advanced. Each exercise comes with its own solution. Besides the exercises, you will find boxes labeled “Discussion,” which contain questions for discussion with your peers or instructor.