Function Level Profiling
Last updated on 2026-01-30 | Edit this page
Estimated time: 40 minutes
Overview
Questions
- When is function level profiling appropriate?
- How can
cProfileandsnakevizbe used to profile a Python program? - How are the outputs from function level profiling interpreted?
Objectives
- execute a Python program via
cProfileto collect profiling information about a Python program’s execution - use
snakevizto visualise profiling information output bycProfile - interpret
snakevizviews, to identify the functions where time is being spent during a program’s execution
Introduction
Software is typically comprised of a hierarchy of function calls, both functions written by the developer and those used from the language’s standard library and third party packages.
Function-level profiling analyses where time is being spent with respect to functions. Typically function-level profiling will calculate the number of times each function is called and the total time spent executing each function, inclusive and exclusive of child function calls.
This allows functions that occupy a disproportionate amount of the total runtime to be quickly identified and investigated.
In this episode we will cover the usage of the function-level
profiler cProfile, how its output can be visualised with
snakeviz and how the output can be interpreted.
What is a Call Stack?
The call stack keeps track of the active hierarchy of function calls and their associated variables.
As a stack it is a last-in first-out (LIFO) data structure.
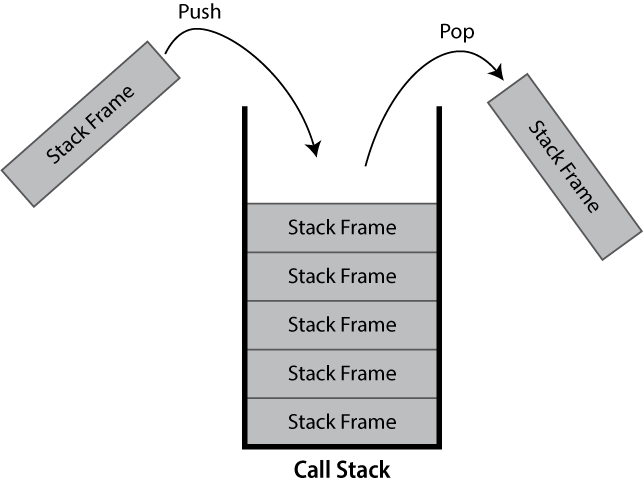
When a function is called, a frame to track its variables and metadata is pushed to the call stack. When that same function finishes and returns, it is popped from the stack and variables local to the function are dropped.
If you’ve ever seen a stack overflow error, this refers to the call stack becoming too large. These are typically caused by recursive algorithms, whereby a function calls itself, that don’t exit early enough.
Within Python the current call-stack can be printed using the core
traceback package, traceback.print_stack()
will print the current call stack.
The below example:
PYTHON
import traceback
def a():
b1()
b2()
def b1():
pass
def b2():
c()
def c():
traceback.print_stack()
a()Here we can see that the printing of the stack trace is called in
c(), which is called by b2(), which is called
by a(), which is called from global scope.
Hence, this prints the following call stack:
OUTPUT
File "C:\call_stack.py", line 13, in <module>
a()
File "C:\call_stack.py", line 5, in a
b2()
File "C:\call_stack.py", line 9, in b2
c()
File "C:\call_stack.py", line 11, in c
traceback.print_stack()The first line states the file and line number where a()
was called from (the last line of code in the file shown). The second
line states that it was the function a() that was called,
this could include its arguments. The third line then repeats this
pattern, stating the line number where b2() was called
inside a(). This continues until the call to
traceback.print_stack() is reached.
You may see stack traces like this when an unhandled exception is thrown by your code.
In this instance the base of the stack has been printed first, other visualisations of call stacks may use the reverse ordering.
cProfile
cProfile
is a function-level profiler provided as part of the Python standard
library.
It can be called directly within your Python code as an imported package, however it’s easier to use its script interface:
For example if you normally run your program as:
You would call cProfile to produce profiling output
out.prof with:
No additional changes to your code are required, it’s really that simple!
If you don’t specify output to file (e.g. remove
-o out.prof from the command), cProfile will
produce output to console similar to that shown below:
OUTPUT
28 function calls in 4.754 seconds
Ordered by: standard name
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 4.754 4.754 worked_example.py:1(<module>)
1 0.000 0.000 1.001 1.001 worked_example.py:13(b_2)
3 0.000 0.000 1.513 0.504 worked_example.py:16(c_1)
3 0.000 0.000 1.238 0.413 worked_example.py:19(c_2)
3 0.000 0.000 0.334 0.111 worked_example.py:23(d_1)
1 0.000 0.000 4.754 4.754 worked_example.py:3(a_1)
3 0.000 0.000 2.751 0.917 worked_example.py:9(b_1)
1 0.000 0.000 4.754 4.754 {built-in method builtins.exec}
11 4.753 0.432 4.753 0.432 {built-in method time.sleep}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}The columns have the following definitions:
| Column | Definition |
|---|---|
ncalls |
The number of times the given function was called. |
tottime |
The total time spent in the given function, excluding child function calls. |
percall |
The average tottime per function call
(tottime/ncalls). |
cumtime |
The total time spent in the given function, including child function calls. |
percall |
The average cumtime per function call
(cumtime/ncalls). |
filename:lineno(function) |
The location of the given function’s definition and its name. |
This output can often exceed the terminal’s buffer length for large
programs and can be unwieldy to parse, so the package
snakeviz is often utilised to provide an interactive
visualisation of the data when exported to file.
snakeviz
It can help to run these examples by running snakeviz
live. For the worked example you may wish to also show the code (e.g. in
split screen).
Demonstrate features such as moving up/down the call-stack by clicking the boxes and changing the depth and cutoff via the dropdown.
Download pre-generated profile reports:
snakeviz example screenshot: files/schelling_out.prof
Worked example: files/snakeviz-worked-example/out.prof
snakeviz
is a web browser based graphical viewer for cProfile output
files.
It is not part of the Python standard library, and therefore must be
installed via pip.
Once installed, you can visualise a cProfile output file
such as out.prof via:
This should open your web browser displaying a page similar to that below.

snakeviz.The icicle diagram displayed by snakeviz represents an
aggregate of the call stack during the execution of the profiled code.
The box which fills the top row represents the root call, filling the
row shows that it occupied 100% of the runtime. The second row holds the
child methods called from the root, with their widths relative to the
proportion of runtime they occupied. This continues with each subsequent
row, however where a method only occupies 50% of the runtime, its
children can only occupy a maximum of that runtime hence the appearance
of “icicles” as each row gets narrower when the overhead of methods with
no further children is accounted for.
By clicking a box within the diagram, it will “zoom” making the selected box the root allowing more detail to be explored. The diagram is limited to 10 rows by default (“Depth”) and methods with a relatively small proportion of the runtime are hidden (“Cutoff”).
As you hover each box, information to the left of the diagram updates specifying the location of the method and for how long it ran.
snakeviz Inside Notebooks
If you’re more familiar with writing Python inside Jupyter notebooks
you can still use snakeviz directly from inside notebooks
using the notebooks “magic” prefix (%) and it will
automatically call cProfile for you.
First snakeviz must be installed and its extension
loaded.
Following this, you can either call %snakeviz to profile
a function defined earlier in the notebook.
Or, you can create a %%snakeviz cell, to profile the
python executed within it.
In both cases, the full snakeviz profile visualisation
will appear as an output within the notebook!
You may wish to right click the top of the output, and select “Disable Scrolling for Outputs” to expand its box if it begins too small.
Worked Example
Demonstrate this!
To more clearly demonstrate how an execution hierarchy maps to the icicle diagram, the below toy example Python script has been implemented.
PYTHON
import time
def a_1():
for i in range(3):
b_1()
time.sleep(1)
b_2()
def b_1():
c_1()
c_2()
def b_2():
time.sleep(1)
def c_1():
time.sleep(0.5)
def c_2():
time.sleep(0.3)
d_1()
def d_1():
time.sleep(0.1)
# Entry Point
a_1()All of the methods except for b_1() call
time.sleep(), this is used to provide synthetic bottlenecks
to create an interesting profile.
-
a_1()callsb_1()x3 andb_2()x1 -
b_1()callsc_1()x1 andc_2()x1 -
c_2()callsd_1()
Follow Along
Download the
Python
source for the example or
cProfile
output file and follow along with the worked example on your own
machine.
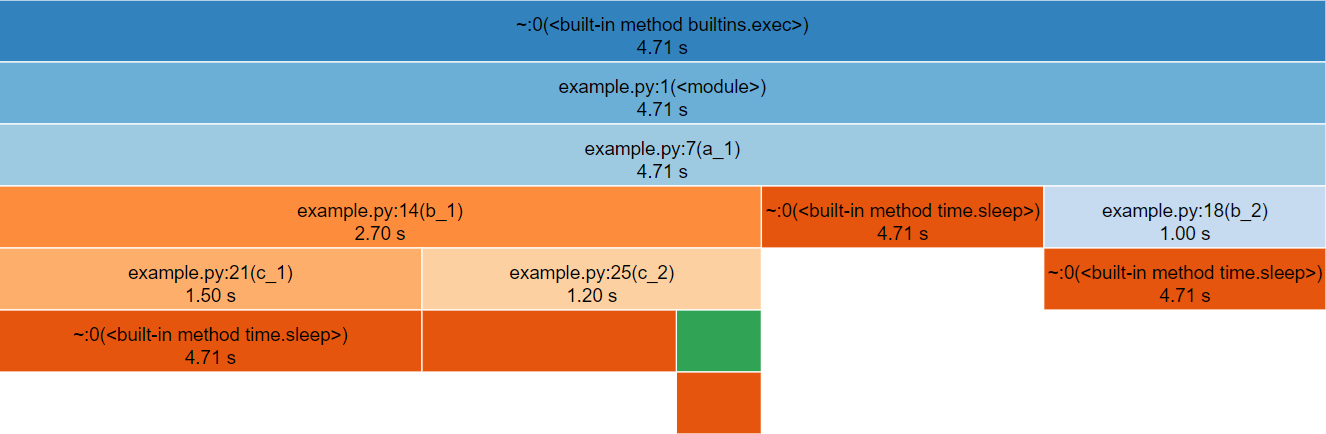
snakeviz for the above Python code.The third row represents a_1(), the only method called
from global scope, therefore the first two rows represent Python’s
internal code for launching our script and can be ignored (by clicking
on the third row).
The row following a_1() is split into three boxes
representing b_1(), time.sleep() and
b_2(). Note that b_1() is called three times,
but only has one box within the icicle diagram. The boxes are ordered
left-to-right according to cumulative time, which happens to be the
order they were first called.
If the box for time.sleep() is hovered it will change
colour along with several other boxes that represent the other locations
that time.sleep() was called from. Note that each of these
boxes display the same duration, the timing statistics collected by
cProfile (and visualised by snakeviz) are
aggregate, so there is no information about individual method calls for
methods which were called multiple times. This does however mean that if
you check the properties to the left of the diagram whilst hovering
time.sleep() you will see a cumulative time of 99%
reported, the overhead of the method calls and for loop is insignificant
in contrast to the time spent sleeping!
Below are the properties shown, the time may differ if you generated the profile yourself.
-
Name:
<built-in method time.sleep> -
Cumulative Time:
4.71 s (99.99 %) -
File:
~ -
Line:
0 - Directory:
As time.sleep() is a core Python method it is displayed
as “built-in method” and doesn’t have a file, line or directory.
If you hover any boxes representing the methods from the above code, you will see file and line properties completed. The directory property remains empty as the profiled code was in the root of the working directory. A profile of a large project with many files across multiple directories will see this filled.
Find the box representing c_2() on the icicle diagram,
its children are unlabelled because they are not wide enough (but they
can still be hovered). Clicking c_2() zooms in the diagram,
showing the children to be time.sleep() and
d_1().
To zoom back out you can either click the top row, which will zoom out one layer, or click “Reset Zoom” on the left-hand side.
In this simple example the execution is fairly evenly balanced between all of the user-defined methods, so there is not a clear hot-spot to investigate.
Below the icicle diagram, there is a table similar to the default
output from cProfile. However, in this case you can sort
the columns by clicking their headers and filter the rows shown by
entering a filename in the search box. This allows built-in methods to
be hidden, which can make it easier to highlight optimisation
priorities.
Notebooks
If you followed along inside a notebook it might look like this:
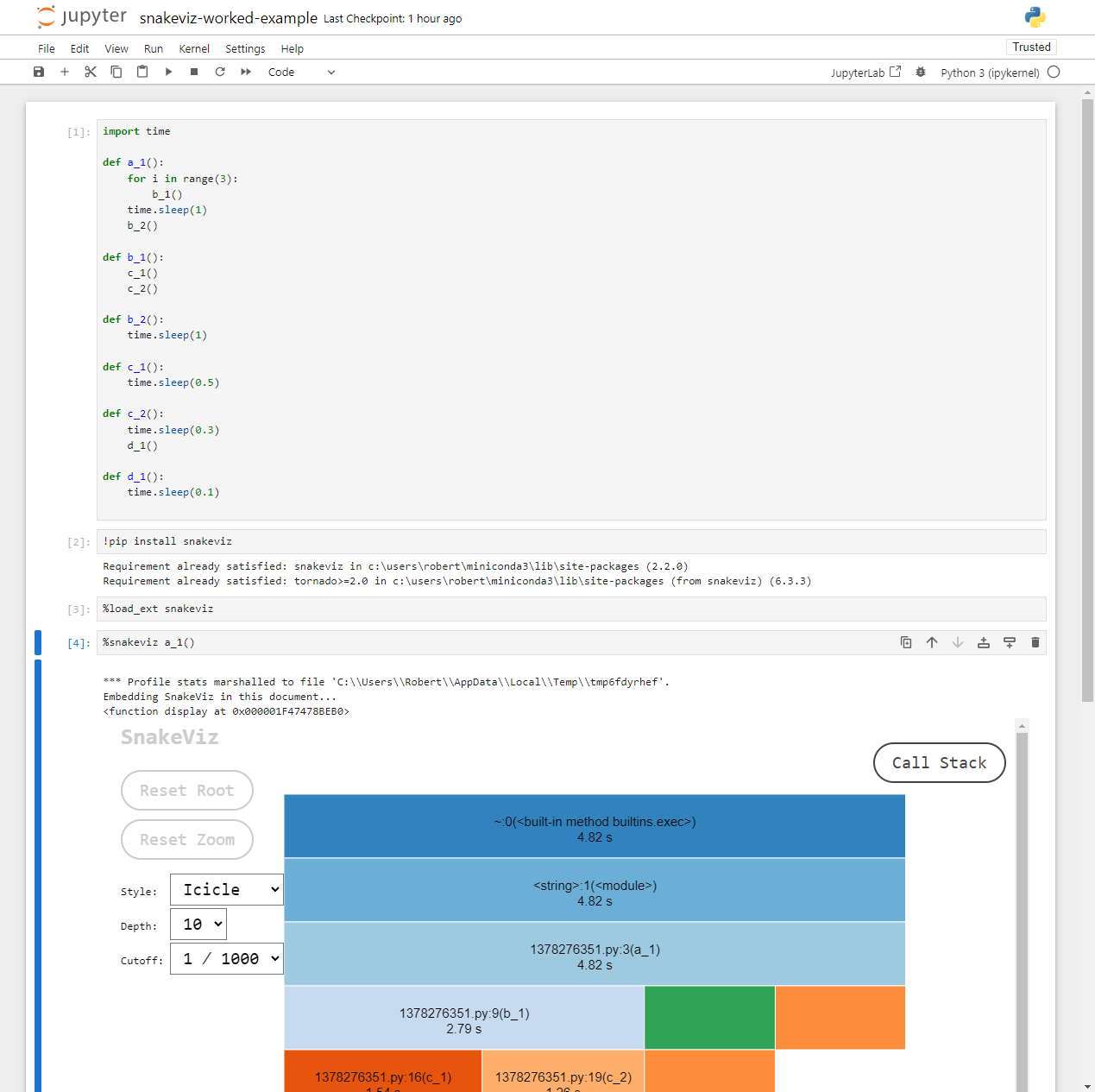
Because notebooks operate by creating temporary Python files, the
filename (shown 1378276351.py above) and line numbers
displayed are not too useful. However, the function names match those
defined in the code and follow the temporary file name in parentheses,
e.g. 1378276351.py:3(a_1),
1378276351.py:9(b_1) refer to the functions
a_1() and b_1() respectively.
Sunburst
snakeviz provides an alternate “Sunburst” visualisation,
accessed via the “Style” drop-down on the left-hand side.
This provides the same information as “Icicle”, however the rows are instead circular with the root method call found at the center.
The sunburst visualisation displays less text on the boxes, so it can be harder to interpret. However, it increases the visibility of boxes further from the root call.
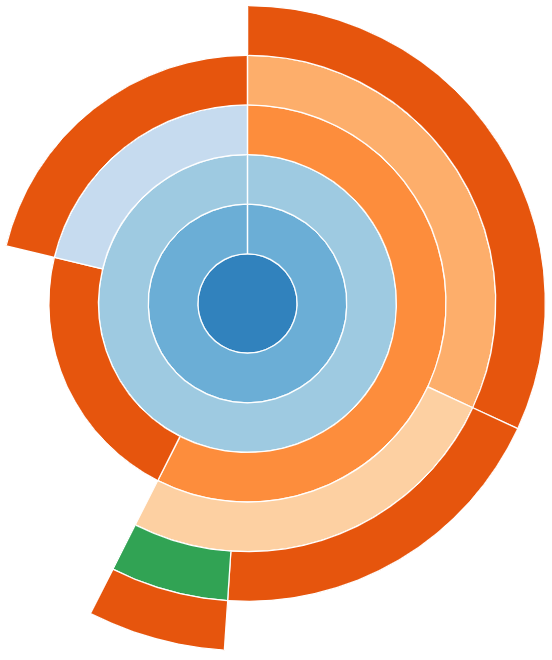
snakeviz for the worked example’s Python code.Exercises
The following exercises allow you to review your understanding of what has been covered in this episode.
Arguments 1-9 passed to travellingsales.py should
execute relatively fast (less than a minute)
This will be slower via the profiler, and is likely to vary on different hardware.
Larger values should be avoided.
Download the set of profiles for arguments 1-10, these can be opened
by passing the directory to snakeviz.
Exercise 1: Travelling Salesperson
Download and profile this Python program, try to locate the function call(s) where the majority of execution time is being spent.
The travelling salesperson problem aims to optimise the route for a scenario where a salesperson is requires to travel between N locations. They wish to travel to each location exactly once, in any order, whilst minimising the total distance travelled.
The provided implementation uses a naive brute-force approach.
The program can be executed via
python travellingsales.py <cities>. The value of
cities should be a positive integer, this algorithm has
poor scaling so larger numbers take significantly longer to run.
- If a hotspot isn’t visible with the argument
1, try increasing the value. - If you think you identified the hotspot with your first profile, try
investigating how the value of
citiesaffects the hotspot within the profile.
The hotspot only becomes visible when an argument of 5
or greater is passed.
You should see that distance() (from
travellingsales.py:11) becomes the largest box (similarly
its parent in the call-stack total_distance()) showing that
it scales poorly with the number of cities. With 5 cities,
distance() has a cumulative time of ~35% the
runtime, this increases to ~60% with 9 cities.
Other boxes within the diagram correspond to the initialisation of imports, or initialisation of cities. These have constant or linear scaling, so their cost barely increases with the number of cities.
This highlights the need to profile a realistic test-case expensive enough that initialisation costs are not the most expensive component.
The default configuration of the Predator Prey model takes around 10 seconds to run, it may be slower on other hardware.
Download the pre-generated cProfile output, this can be
opened with snakeviz to save waiting for the profiler.
Exercise 2: Predator Prey
Download and profile the Python predator prey model, try to locate the function call(s) where the majority of execution time is being spent
This exercise uses the packages numpy and
matplotlib, they can be installed via
pip install numpy matplotlib.
The predator prey model is a simple agent-based model of population dynamics. Predators and prey co-exist in a common environment and compete over finite resources.
The three agents; predators, prey and grass exist in a two dimensional grid. Predators eat prey, prey eat grass. The size of each population changes over time. Depending on the parameters of the model, the populations may oscillate, grow or collapse due to the availability of their food source.
The program can be executed via
python predprey.py <steps>. The value of
steps for a full run is 250, however a full run may not be
necessary to find the bottlenecks.
When the model finishes it outputs a graph of the three populations
predprey_out.png.
It should be clear from the profile that the method
Grass::eaten() (from predprey.py:278) occupies
the majority of the runtime.
From the table below the Icicle diagram, we can see that it was called 1,250,000 times.
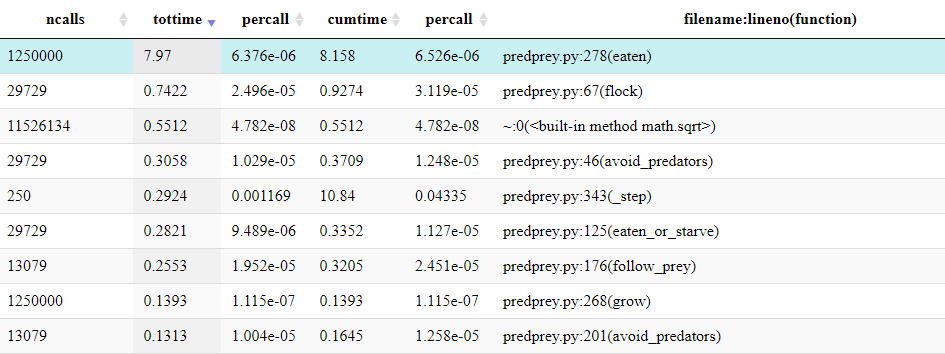
If the table is ordered by ncalls, it can be identified
as the joint 4th most called method and 2nd most called method from
predprey.py.
If you checked predprey_out.png (shown below), you
should notice that there are significantly more Grass
agents than Predators or Prey.
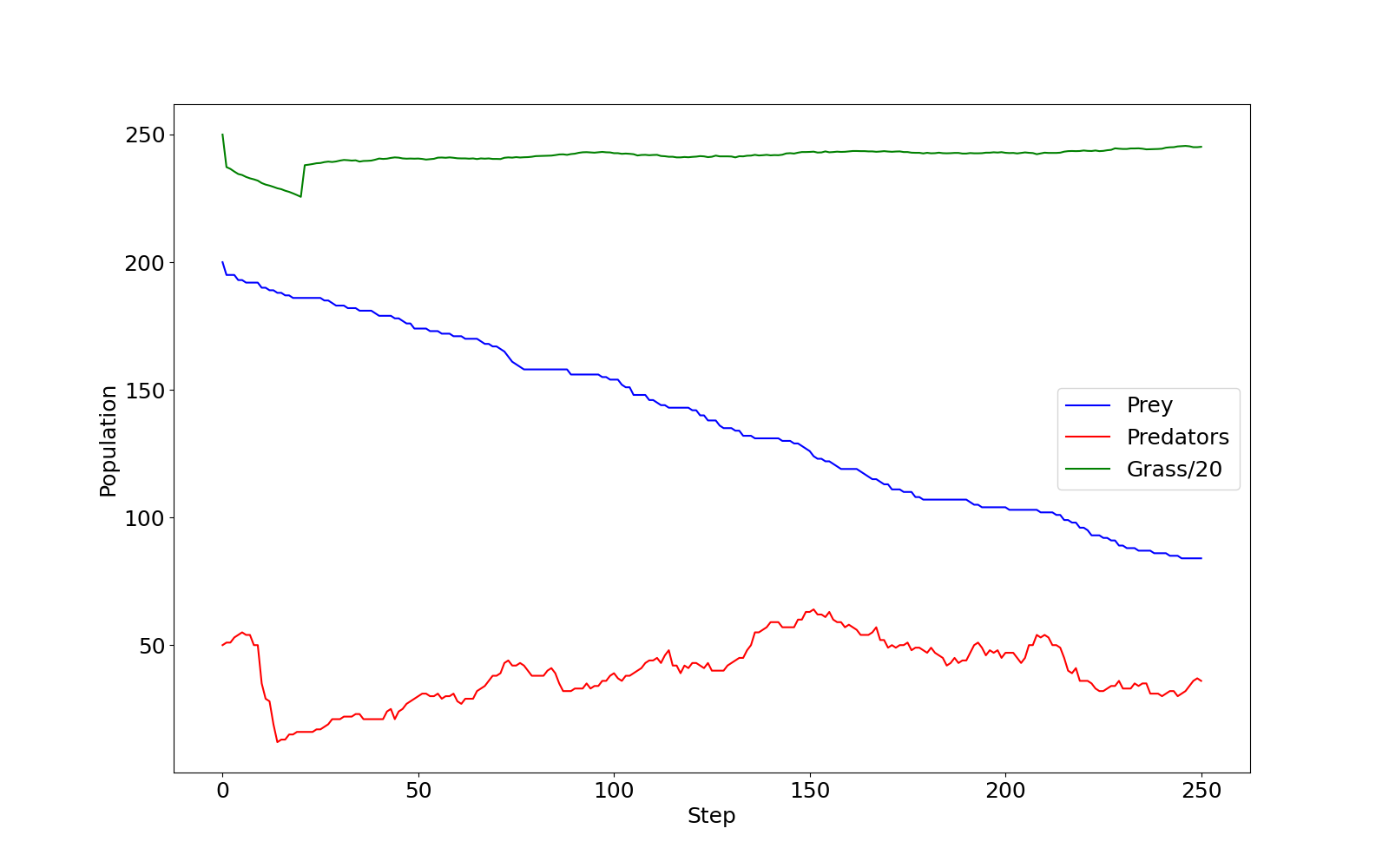
predprey_out.png as produced by the
default configuration of predprey.py.Similarly, the Grass::eaten() has a percall
time is inline with other agent functions such as
Prey::flock() (from predprey.py:67).
Maybe we could investigate this further with line profiling!
You may have noticed many iciles on the right hand of the
diagram, these primarily correspond to the import of
matplotlib which is relatively expensive!
- A python program can be function level profiled with
cProfileviapython -m cProfile -o <output file> <script name> <arguments>. - The output file from
cProfilecan be visualised withsnakevizviapython -m snakeviz <output file>. - Function level profiling output displays the nested call hierarchy, listing both the cumulative and total minus sub functions time.
