Astronomy Workflow: Idea to Implementation
Overview
Teaching: 30 min
Exercises: 30 minQuestions
How do I develop a workflow from scratch?
Objectives
Create a workflow from scratch and see it run.
The idea
We wish to create a workflow which will process observing data from some telescope, find candidate pulsars, and then run a ML on these candidates to label them as confirmed or rejected. Humans will then sort through the confirmed list to come up with a final decision on which are real and which are not.
The high level workflow therefore looks like this:
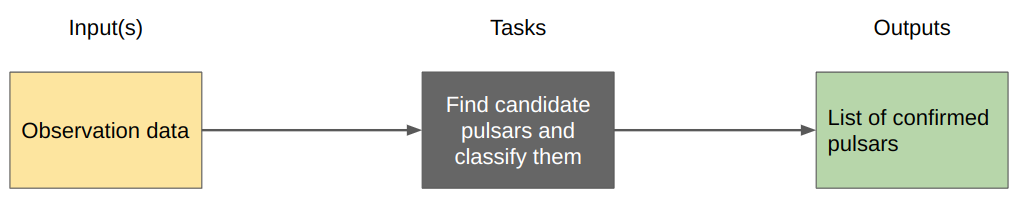
As we think more about how the processing will be done we come up with some intermediate steps for the tasks:
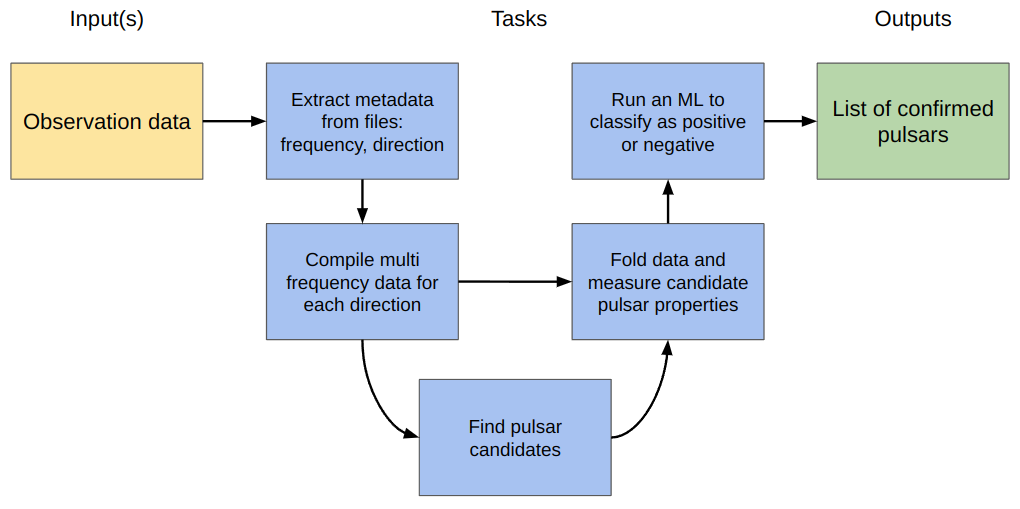
In this final iteration we have indicated with arrows the dependencies of each task, but also where information needs to be passed. For example the “fold data” task needs the multi-frequency data from the “compile” task, as well as the candidate pulsar details from the “find” task in order to do a more detailed measurement of the properties.
The process
Step 1: Initial channel
Within the example files,
is a set of files in the observations directory with names like obs_[1-9].dat.
The meta-data for each file is stored in the file header.
In our example we don’t have any actual data in the files, just meta-data, and the files look like this:
obs_1.dat
#freq: 10 MHz #date: 2022-11-15 #point: 10deg_10deg
Our initial channel will be a list of all the files in the observations directory that look like obs_*.dat.
We will leave the data directory as a user parameter with a default value of observations.
We create our channel as follows:
params.data_dir = "observations"
all_obs = Channel.fromPath("${params.data_dir}/*.dat")
Step 2: Extracting meta-data
The first step in our workflow is to extract the metadata from the files.
For this we define a process called get_meta.
Since this will be the first process in the workflow we set it up to accept input as single files, and it will return a tuple of the frequency and pointing direction for the given file, as well as the file itself.
process get_meta {
// Convert an input file into a stream with meta data
// output will be [frequency, point, file]
input:
path(obs)
output:
tuple (env(FREQ), env(POINT), path(obs))
script:
"""
FREQ=\$(grep -e "#freq:" ${obs} | awk '{print \$2}')
POINT=\$(grep -e "#point:" ${obs} | awk '{print \$2}')
"""
}
Things to note above:
- We can return variables from our (bash) scripts by saving them as environment variables, and telling NextFlow that the output type is env.
- We have escaped bash
$with a\. - We don’t modify or create any files here, we just extract variables, so the script will take almost no time to run.
To test how we are going so far we can create a stub of a workflow in the same file with some .view() commands on the various channels that we are using.
workflow {
all_obs.view() // view channel
get_meta(all_obs) // run process
get_meta.out.view() // view the channel generated by the output
}
We run the above with nextflow run astro_wf.nf and it gives us the following:
$ nextflow run astro_wf.nf
N E X T F L O W ~ version 22.10.1
Launching `astro_wf.nf` [zen_einstein] DSL2 - revision: 5338dcdf52
executor > local (9)
[17/bcae92] process > get_meta (9) [100%] 9 of 9 ✔
/data/some/dir/observations/obs_4.dat
/data/some/dir/observations/obs_2.dat
/data/some/dir/observations/obs_9.dat
/data/some/dir/observations/obs_3.dat
/data/some/dir/observations/obs_7.dat
/data/some/dir/observations/obs_8.dat
/data/some/dir/observations/obs_1.dat
/data/some/dir/observations/obs_6.dat
/data/some/dir/observations/obs_5.dat
[30, 20deg_10deg, /data/some/dir/work/3d/e3663a45357b9818a547c2908701da/obs_6.dat]
[10, 20deg_10deg, /data/some/dir/work/24/69ad9546cacb3ff01b6199977ab80d/obs_4.dat]
[30, 30deg_10deg, /data/some/dir/work/e5/44d3d5698dc36454861601e4665a32/obs_9.dat]
[20, 30deg_10deg, /data/some/dir/work/bd/e29f1747bd01c8cc8f2278f29f1b17/obs_8.dat]
[20, 10deg_10deg, /data/some/dir/work/33/a37ad4d85131c77467126e0d361667/obs_2.dat]
[30, 10deg_10deg, /data/some/dir/work/0f/22afa02fd765ca828341a6aa91630d/obs_3.dat]
[10, 10deg_10deg, /data/some/dir/work/f4/c5618dd7b5aa66b23c3ce4c1ddc1d3/obs_1.dat]
[10, 30deg_10deg, /data/some/dir/work/19/8411e3effcb8eb6b13d1ae450b3256/obs_7.dat]
[20, 20deg_10deg, /data/some/dir/work/17/bcae92db0ed291c969217d979374a8/obs_5.dat]
We can see from the above that the input files are obs_?.dat, and that the get_meta process then turns these into the required tuple of [freq, pointing, file].
Note that when the files are passed to get_meta they are symlinked to a new work folder so the file names remain the same but the directory path is updated.
At this point we can see that the get_meta is working as intended so we can move on.
Step 3: Compiling multi-frequency data
The next step in the workflow is to combine all the files that share the same pointing direction. This will result a multi-frequency data set/file per pointing direction.
There are two things that we need to do here.
The first is to take the output of the get_meta process and group it so that all the files that share a common pointing are now within a single tuple.
The second is to create a new process that will combine the data sets.
The channel that comes from get_meta is shown above and you can see that it has one item per [freq, pointing, filename], with some of the freq and pointing overlapping.
What we want instead is [[freq1, freq2, ...], pointing, [datafile1, datafile2, ...]], which means that instead of 9 items in the channel we have just three (corresponding to the three pointing directions).
To achieve this goal we use the groupTuple method on the get_meta.out channel.
By default groupTuple will use the first item in a tuple as the key, but we want it to use the pointing which is the second item (index=1).
Therefore we modify our workflow to create the new channel called same_pointing, and remove the .view() on the all_obs channel.
workflow {
// create metadata
get_meta( all_obs )
// collect all the files that have the same pointing
get_meta.out.view()
same_pointing = get_meta.out.groupTuple( by: 1 ).view()
}
We now re-run the workflow to make sure that we are getting the required channel ‘shape’.
$ nextflow run astro_wf.nf
N E X T F L O W ~ version 22.10.1
Launching `astro_wf.nf` [fabulous_woese] DSL2 - revision: d4bc8976c3
executor > local (9)
[8f/01c418] process > get_meta (9) [100%] 9 of 9 ✔
[30, 20deg_10deg, work/59/7cd29865c2ef0d56cd3ca8a3b824f6/obs_6.dat]
executor > local (9)
[8f/01c418] process > get_meta (9) [100%] 9 of 9 ✔
[30, 20deg_10deg, work/59/7cd29865c2ef0d56cd3ca8a3b824f6/obs_6.dat]
[20, 10deg_10deg, work/cb/b4d7c2370b9fa1babba1dc4f2f1334/obs_2.dat]
[10, 30deg_10deg, work/03/a53409f45b318ba49990b8a33dc29c/obs_7.dat]
[30, 10deg_10deg, work/fe/039db662fe5bd31024c745412791d3/obs_3.dat]
[20, 30deg_10deg, work/26/760d76cf79bf2b970cfaf8d67ea797/obs_8.dat]
[10, 10deg_10deg, work/0d/76acb6d5e7b507af1ab4fe018f500f/obs_1.dat]
[30, 30deg_10deg, work/ee/596bb5bad24c60ab84c7efc74b1bd0/obs_9.dat]
[10, 20deg_10deg, work/42/cc5d006b6a514928ee17333de95e02/obs_4.dat]
[20, 20deg_10deg, work/8f/01c4180f254a1900dfe5135ccfbc50/obs_5.dat]
[[30, 10, 20], 20deg_10deg, [work/59/7cd29865c2ef0d56cd3ca8a3b824f6/obs_6.dat, work/42/cc5d006b6a514928ee17333de95e02/obs_4.dat, work/8f/01c4180f254a1900dfe5135ccfbc50/obs_5.dat]]
[[20, 30, 10], 10deg_10deg, [work/cb/b4d7c2370b9fa1babba1dc4f2f1334/obs_2.dat, work/fe/039db662fe5bd31024c745412791d3/obs_3.dat, work/0d/76acb6d5e7b507af1ab4fe018f500f/obs_1.dat]]
[[10, 20, 30], 30deg_10deg, [work/03/a53409f45b318ba49990b8a33dc29c/obs_7.dat, work/26/760d76cf79bf2b970cfaf8d67ea797/obs_8.dat, work/ee/596bb5bad24c60ab84c7efc74b1bd0/obs_9.dat]]
The order in which the files are being grouped is different for each pointing direction. Note, however that the mapping between the frequency and filenames are still correct (eg. obs_1.dat is paired with 10 in pointing 10deg_10deg).
Now that the channel is working we can set up a process to join the data for each pointing direction.
We set the input to be a tuple with the same shape as the input channel.
All process inputs can be a single value/file or a list of values/files.
This flexibility means that this process would still work if we input more than three files to this process.
In this example, the variable freqs will be a list of values, and obs will be a list of file objects.
Our dummy script is just going to concatenate all the observations into a single file with a name that is like obs_f1_f2_f3.dat.
Of course in reality you’ll have some better way of doing this.
Finally, our output is going to be the pointing direction and the file that was just generated.
process combine_frequencies {
// Combine the files so the output has a single pointing with all the frequency information
input:
tuple (val(freqs), val(point), path(obs))
output:
tuple (val(point), path("obs*dat"))
script:
"""
cat ${obs} > obs_${freqs.join("_")}.dat
"""
}
Note that ${obs} and ${freqs} are both lists so we have access to the .join() function.
Let’s now update our workflow description to include the above, and modify the workflow section to show what is going into and coming out of the combine_frequencies process.
workflow {
// create metadata
get_meta( all_obs )
// collect all the files that have the same pointing
same_pointing = get_meta.out.groupTuple( by: 1 ).view()
// Combine the frequencies so you have a single file with all frequencies
combine_frequencies( same_pointing )
combine_frequencies.out.view()
}
Our output is now:
$ nextflow run astro_wf.nf
N E X T F L O W ~ version 22.10.1
Launching `astro_wf.nf` [romantic_rosalind] DSL2 - revision: cdeceec8f1
executor > local (12)
[c4/19d3d3] process > get_meta (9) [100%] 9 of 9 ✔
[17/f20f77] process > combine_frequencies (1) [100%] 3 of 3 ✔
[[30, 20, 10], 10deg_10deg, [/work/4b/89cc7d4a29c63036def3945a00dfdc/obs_3.dat, /work/f5/2c181e756aacb93620edf61db87781/obs_2.dat, /work/d7/8e86a29a4046159431e7021883ebb8/obs_1.dat]]
[[30, 20, 10], 30deg_10deg, [/work/d7/2ff98b2b97bf9535dbdc47bd90a9a9/obs_9.dat, /work/07/f118faf09efc8738034a4a944392a6/obs_8.dat, /work/77/d9299b3367c8ccbbf47e425abc6133/obs_7.dat]]
[[30, 10, 20], 20deg_10deg, [/work/42/3d0cd8f5f1a3fc9a088f4eade1fcb2/obs_6.dat, /work/4e/02d9f520d266ea5cb5160db6297105/obs_4.dat, /work/c4/19d3d3528fe39a03d0e673969d7239/obs_5.dat]]
[30deg_10deg, /work/52/6bded1ba8fabfaedb5fc1d8bab1ab5/obs_30_20_10.dat]
[20deg_10deg, /work/ac/525ce379788357bd0d996e4cecc444/obs_30_10_20.dat]
[10deg_10deg, /work/17/f20f775aa7866c31077070f1a8715e/obs_30_20_10.dat]
Let’s have a look at how NextFlow is doing the variable replacement for lists in the combine_frequencies process.
To do this we open the working directory for the 10deg_10deg pointing (/work/17/f20f775aa7866c31077070f1a8715e/) and look at the .command.sh file.
#!/bin/bash -ue
cat obs_3.dat obs_2.dat obs_1.dat > obs_30_20_10.dat
Here you can see that ${obs} was expended to be obs_3.dat obs_2.dat obs_1.dat, and our tuple of frequencies [30,20,10] was replaced with 30_20_10 thanks to the .join("_") operator.
Step 4: Find candidates
We now have a channel with items like: [pointing, combined data file].
This is in the format that we want for searching for candidates so we don’t need to mutate the channel at all, and can go straight to making the new process.
In our dummy script we are just making a random number of candidates using a bash script (the real processing is commented out).
Since this script uses a lot of bash variables we can avoid escaping the $ by creating a shell instead of a script section (remember to use ''' not """).
Since the number of candidates that are found is random, and possibly zero, we indicate that the output is optional (remember to use path as file does not have this option).
This means that if the process runs and there are no files matching cand*dat then no items are emitted into the output channel but the process still completes without failure.
process find_candidates {
// Use a periodicity search to find events with significance above 6sigma
input:
tuple (val(point), path(obs))
output:
tuple (val(point), path("cand*dat"), optional: true)
shell:
'''
#./find_periodic_cands.sh !{obs}
# Random number from 0-3
ncand=$(( $RANDOM % 4 ))
echo $ncand
for i in $(seq ${ncand}); do
touch cand_${i}.dat
done
'''
}
Again we modify our workflow section so that we can see just the input/output for this process as we test and debug our workflow.
workflow {
// create metadata
get_meta( all_obs )
// collect all the files that have the same pointing
same_pointing = get_meta.out.groupTuple( by: 1 )
// Combine the frequencies so you have a single file with all frequencies
combine_frequencies( same_pointing )
// Look for periodic signals with an fft
combine_frequencies.out.view()
find_candidates( combine_frequencies.out )
find_candidates.out.view()
}
When I run I get the following output. Your output will differ because the number of candidates is random. Here you can see that I got zero candidates for the 20deg_10_deg pointing, so there are just two items in my output channel.
$ nextflow run astro_wf.nf
N E X T F L O W ~ version 22.10.1
Launching `astro_wf.nf` [fervent_shockley] DSL2 - revision: 7a02379938
executor > local (15)
[ac/dfbd2c] process > get_meta (2) [100%] 9 of 9 ✔
[62/9e9dec] process > combine_frequencies (2) [100%] 3 of 3 ✔
[7e/d651a0] process > find_candidates (1) [100%] 3 of 3 ✔
[20deg_10deg, /work/e4/f2898b75b400ed0fda4af3a53ced16/obs_10_30_20.dat]
[10deg_10deg, /work/9b/7b5d6f6c6e4c139c46bf23330756ff/obs_30_10_20.dat]
[30deg_10deg, /work/62/9e9decdb609c0cefd36586412f25f9/obs_30_10_20.dat]
[10deg_10deg, [/work/e3/3983bf3b27049a2fbd281786dab0ed/cand_1.dat, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_2.dat]]
[30deg_10deg, [/work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_1.dat, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_2.dat, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_3.dat]]
Step 5: Measure candidate properties
This next process requires that we combine two input channels.
We need the multi frequency data files which are provided by the combine_frequencies process, as well as the corresponding pulsar candidates provided by the find_candidates process.
Recall the format for these two channels:
combine_frequencies.outlooks like[pointing, mf_file]find_candidates.outlooks like[pointing, [candidate_file1, ...] ]
We want our new process to work on a single candidate at a time so that if there are two candidates for a single input file, we’ll run the process two times.
Thus we want the input channel to look like [pointing, mf_file, candidate_file].
We could just use cross to join the two channels together using pointing as the key, however it will give only one item per pointing, rather than one item per candidate.
Therefore we need to modify the find_candidates.out channel so that it has a single [pointing, candidate_file] pair per candidate.
The NextFlow operator for this is transpose and we use it like this:
workflow {
// create metadata
get_meta( all_obs )
// collect all the files that have the same pointing
same_pointing = get_meta.out.groupTuple( by: 1 )
// Combine the frequencies so you have a single file with all frequencies
combine_frequencies( same_pointing )
// Look for periodic signals with an fft
find_candidates( combine_frequencies.out )
find_candidates.out.view()
// tranpose will "flatten" the cands so they have the format [ key, cand_file ]
flattened_cands = find_candidates.out.transpose()
flattened_cands.view()
}
This time I run the workflow using the -resume option so that I pick up from my previous run, and will get the same candidates as before.
$ nextflow run astro_wf.nf -resume
N E X T F L O W ~ version 22.10.1
Launching `astro_wf.nf` [backstabbing_murdock] DSL2 - revision: fc5d3ca357
[4e/b890fe] process > get_meta (9) [100%] 9 of 9, cached: 9 ✔
[9b/7b5d6f] process > combine_frequencies (3) [100%] 3 of 3, cached: 3 ✔
[e3/3983bf] process > find_candidates (3) [100%] 3 of 3, cached: 3 ✔
[30deg_10deg, [/work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_1.dat, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_2.dat, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_3.dat]]
[10deg_10deg, [/work/e3/3983bf3b27049a2fbd281786dab0ed/cand_1.dat, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_2.dat]]
[30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_1.dat]
[30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_2.dat]
[30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_3.dat]
[10deg_10deg, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_1.dat]
[10deg_10deg, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_2.dat]
Now we are in a position to join the two channels together with cross using the pointing as the key:
workflow {
// create metadata
get_meta( all_obs )
// collect all the files that have the same pointing
same_pointing = get_meta.out.groupTuple( by: 1 )
// Combine the frequencies so you have a single file with all frequencies
combine_frequencies( same_pointing )
// Look for periodic signals with an fft
find_candidates( combine_frequencies.out )
// transpose will "flatten" the cands so they have the format [ key, cand_file ]
flattened_cands = find_candidates.out.transpose()
flattened_cands.view()
combine_frequencies.out.view()
// For each candidate file pair it with observation file
cand_obs_pairs = combine_frequencies.out.cross(flattened_cands)
cand_obs_pairs.view()
}
The output can get mixed up since the processes don’t execute in serial, so below I have reordered the outputs and added some comments (##) to show the different channels.
$ nextflow run astro_wf.nf -resume
N E X T F L O W ~ version 22.10.1
Launching `astro_wf.nf` [festering_minsky] DSL2 - revision: 213dbb16a9
[4e/b890fe] process > get_meta (9) [100%] 9 of 9, cached: 9 ✔
[9b/7b5d6f] process > combine_frequencies (1) [100%] 3 of 3, cached: 3 ✔
[e3/3983bf] process > find_candidates (2) [100%] 3 of 3, cached: 3 ✔
## These are from flattened_cands = find_candidates.out.transpose()
[30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_1.dat]
[30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_2.dat]
[10deg_10deg, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_2.dat]
[30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_3.dat]
[10deg_10deg, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_1.dat]
## These are from combine_frequencies.out
[30deg_10deg, /work/62/9e9decdb609c0cefd36586412f25f9/obs_30_10_20.dat]
[10deg_10deg, /work/9b/7b5d6f6c6e4c139c46bf23330756ff/obs_30_10_20.dat]
[30deg_10deg, /work/62/9e9decdb609c0cefd36586412f25f9/obs_30_10_20.dat]
[10deg_10deg, /work/9b/7b5d6f6c6e4c139c46bf23330756ff/obs_30_10_20.dat]
[20deg_10deg, /work/e4/f2898b75b400ed0fda4af3a53ced16/obs_10_30_20.dat]
## These are cand_obs_pairs = combine_frequencies.out.cross(flattened_cands)
[[30deg_10deg, /work/62/9e9decdb609c0cefd36586412f25f9/obs_30_10_20.dat], [30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_2.dat]]
[[30deg_10deg, /work/62/9e9decdb609c0cefd36586412f25f9/obs_30_10_20.dat], [30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_3.dat]]
[[10deg_10deg, /work/9b/7b5d6f6c6e4c139c46bf23330756ff/obs_30_10_20.dat], [10deg_10deg, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_1.dat]]
[[10deg_10deg, /work/9b/7b5d6f6c6e4c139c46bf23330756ff/obs_30_10_20.dat], [10deg_10deg, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_2.dat]]
[[30deg_10deg, /work/62/9e9decdb609c0cefd36586412f25f9/obs_30_10_20.dat], [30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_1.dat]]
This is almost what we want.
Note that we have a duplicated key (pointing) in the output from the cross operator.
We could make a compilcated input for our new process, but it’s better to just manipulate the channel to remove the key and simplify the tuple.
The relevant part of our workflow is now:
// For each candidate file pair it with observation file
cand_obs_pairs = combine_frequencies.out.cross(flattened_cands)
//reformat to remove redundant key
.map{ [ it[0][0], it[0][1], it[1][1] ] }
// [ pointing, obs_file, candidate_file ]
cand_obs_pairs.view()
And when we run we get the following:
$ nextflow run astro_wf.nf -resume
N E X T F L O W ~ version 22.10.1
Launching `astro_wf.nf` [naughty_ramanujan] DSL2 - revision: a493dd4e99
[4e/b890fe] process > get_meta (9) [100%] 9 of 9, cached: 9 ✔
[e4/f2898b] process > combine_frequencies (1) [100%] 3 of 3, cached: 3 ✔
[e3/3983bf] process > find_candidates (2) [100%] 3 of 3, cached: 3 ✔
## These are from flattened_cands = find_candidates.out.transpose()
[30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_1.dat]
[30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_2.dat]
[30deg_10deg, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_3.dat]
[10deg_10deg, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_1.dat]
[10deg_10deg, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_2.dat]
## These are from combine_frequencies.out
[20deg_10deg, /work/e4/f2898b75b400ed0fda4af3a53ced16/obs_10_30_20.dat]
[10deg_10deg, /work/9b/7b5d6f6c6e4c139c46bf23330756ff/obs_30_10_20.dat]
[30deg_10deg, /work/62/9e9decdb609c0cefd36586412f25f9/obs_30_10_20.dat]
## These are cand_obs_pairs = combine_frequencies.out.cross(flattened_cands).map{ [ it[0][0], it[0][1], it[1][1] ] }
[30deg_10deg, /work/62/9e9decdb609c0cefd36586412f25f9/obs_30_10_20.dat, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_1.dat]
[30deg_10deg, /work/62/9e9decdb609c0cefd36586412f25f9/obs_30_10_20.dat, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_2.dat]
[30deg_10deg, /work/62/9e9decdb609c0cefd36586412f25f9/obs_30_10_20.dat, /work/0a/039acf28cb01c737de9cd2f75c1cc8/cand_3.dat]
[10deg_10deg, /work/9b/7b5d6f6c6e4c139c46bf23330756ff/obs_30_10_20.dat, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_1.dat]
[10deg_10deg, /work/9b/7b5d6f6c6e4c139c46bf23330756ff/obs_30_10_20.dat, /work/e3/3983bf3b27049a2fbd281786dab0ed/cand_2.dat]
We now make another dummy process to work on the candidates.
The resulting .dat file will normally contain some information about the candidate but for now it’s an empty file.
Note that even though we can predict the name of the outfile, we can use *dat to capture whatever is generated.
By default NextFlow will not copy files that are inputs (eg cand_1.dat or obs_30_10_20.dat) to the output so our glob approach is safe here.
If we wanted the inputs to be included there is an option for that (includeInputs: true).
process fold_cands {
// Fold the candidates on the given period and measure properties
// for example: SNR, DM, p, pdot, intensity
input:
tuple (val(point), path(obs), path(cand))
output:
tuple (val(point), path("*dat"))
script:
"""
touch ${point}_${cand.baseName}.dat
"""
}
Now we can update our workflow to run this process and view the output channel.
workflow {
// create metadata
get_meta( all_obs )
// collect all the files that have the same pointing
same_pointing = get_meta.out.groupTuple( by: 1 )
// Combine the frequencies so you have a single file with all frequencies
combine_frequencies( same_pointing )
// Look for periodic signals with an fft
find_candidates( combine_frequencies.out )
//find_candidates.out.view()
// transpose will "flatten" the cands so they have the format [ key, cand_file ]
flattened_cands = find_candidates.out.transpose()
// For each candidate file pair it with observation file
cand_obs_pairs = combine_frequencies.out.cross(flattened_cands)
//reformat to remove redundant key
.map{ [ it[0][0], it[0][1], it[1][1] ] }
// [ pointing, obs_file, candidate_file ]
// Process the candidate
fold_cands( cand_obs_pairs )
fold_cands.out.view()
}
Our output is looking a little more tidy this time.
$ nextflow run astro_wf.nf -resume
N E X T F L O W ~ version 22.10.1
Launching `astro_wf.nf` [stupefied_watson] DSL2 - revision: 762707769d
executor > local (5)
[4e/b890fe] process > get_meta (9) [100%] 9 of 9, cached: 9 ✔
[62/9e9dec] process > combine_frequencies (1) [100%] 3 of 3, cached: 3 ✔
[0a/039acf] process > find_candidates (3) [100%] 3 of 3, cached: 3 ✔
[b0/0f1f30] process > fold_cands (2) [100%] 5 of 5 ✔
[30deg_10deg, /work/21/f0710dda186ddfe6212becc1197413/30deg_10deg_cand_2.dat]
[30deg_10deg, /work/c0/5c1866812c6be0f5ddd212b25e122b/30deg_10deg_cand_1.dat]
[30deg_10deg, /work/65/7124a1e4bacaf3256f5f0a1d9022f3/30deg_10deg_cand_3.dat]
[10deg_10deg, /work/94/314719324a38d621893efbe315ac2a/10deg_10deg_cand_1.dat]
[10deg_10deg, /work/b0/0f1f3005842f76d8c1a19e6c0fd004/10deg_10deg_cand_2.dat]
Step 6: Run some ML on the folded candidates
The output channel from the fold_cands is now nice and simple and it contains all the information we need to run our ML process.
Again, we are using a bash script instead of the actual ML program, so we’ll use a shell.
Note also that we now have two outputs that we are tracking, that they are now directories, and that both are optional.
Furthermore, this is the last process in our workflow so we add a new directive to the process called publishDir which tells NextFlow that we want to copy the outputs from this process to a different directory (not in the work directory).
process ML_thing {
// apply a machine learning algorithm to take the folded data and predict
// real (positve) or fake (negative) candidates
publishDir "cands/", mode: 'copy'
input:
tuple(val(point), path(candidates))
output:
path("positive/*"), optional: true
path("negative/*"), optional: true
shell:
'''
mkdir positive
mkdir negative
for f in !{candidates}; do
if [ $(( $RANDOM % 2 )) == 0 ]; then
mv $f positive/
else
mv $f negative/
fi
done
'''
}
We can tack this onto the end of the workflow and run!
workflow {
// create metadata
get_meta( all_obs )
// collect all the files that have the same pointing
same_pointing = get_meta.out.groupTuple( by: 1 )
// Combine the frequencies so you have a single file with all frequencies
combine_frequencies( same_pointing )
// Look for periodic signals with an fft
find_candidates( combine_frequencies.out )
//find_candidates.out.view()
// transpose will "flatten" the cands so they have the format [ key, cand_file ]
flattened_cands = find_candidates.out.transpose()
// For each candidate file pair it with observation file
cand_obs_pairs = combine_frequencies.out.cross(flattened_cands)
//reformat to remove redundant key
.map{ [ it[0][0], it[0][1], it[1][1] ] }
// [ pointing, obs_file, candidate_file ]
// Process the candidate
fold_cands( cand_obs_pairs )
// // Put the candidates through ML
ML_thing( fold_cands.out )
}
$ nextflow run astro_wf.nf -resume
N E X T F L O W ~ version 22.10.1
Launching `astro_wf.nf` [mad_swartz] DSL2 - revision: 2c5bab1faf
executor > local (5)
[4e/b890fe] process > get_meta (9) [100%] 9 of 9, cached: 9 ✔
[e4/f2898b] process > combine_frequencies (3) [100%] 3 of 3, cached: 3 ✔
[e3/3983bf] process > find_candidates (3) [100%] 3 of 3, cached: 3 ✔
[c0/5c1866] process > fold_cands (3) [100%] 5 of 5, cached: 5 ✔
[61/0371b5] process > ML_thing (5) [100%] 5 of 5 ✔
We have removed all the .view() commands so the output here is much nicer to read.
If we want to see our results we look in the direcory cands
$ tree cands
cands
├── negative
│ ├── 10deg_10deg_cand_1.dat
│ ├── 10deg_10deg_cand_2.dat
│ ├── 20deg_10deg_cand_1.dat
│ ├── 20deg_10deg_cand_2.dat
│ ├── 20deg_10deg_cand_3.dat
│ ├── 30deg_10deg_cand_1.dat
│ ├── 30deg_10deg_cand_2.dat
│ └── 30deg_10deg_cand_3.dat
└── positive
├── 10deg_10deg_cand_1.dat
├── 10deg_10deg_cand_3.dat
├── 20deg_10deg_cand_1.dat
├── 20deg_10deg_cand_2.dat
├── 30deg_10deg_cand_1.dat
├── 30deg_10deg_cand_2.dat
└── 30deg_10deg_cand_3.dat
2 directories, 15 files
Viewing workflow meta-data
Create a .config file
This will create a bunch of useful analysis for your pipeline run when it completes. See next lesson for more about configuration files.
nextflow.config
// turn on all the juicy logging trace.enabled = true timeline.enabled = true report.enabled = true dag { enabled = true file='dag.png' }
If we then rerun our workflow without the -resume option, we’ll get the following files in our directory:
timeline-<date>-<timestamp>.htmlreport-<date>-<timestamp>.htmltrace-<date>-<timestamp>.txtdag.png(ordag.dotif you don’t have graphviz installed)
The first thing we can look at is the dag.png file which is a directed analytic graph of our workflow.
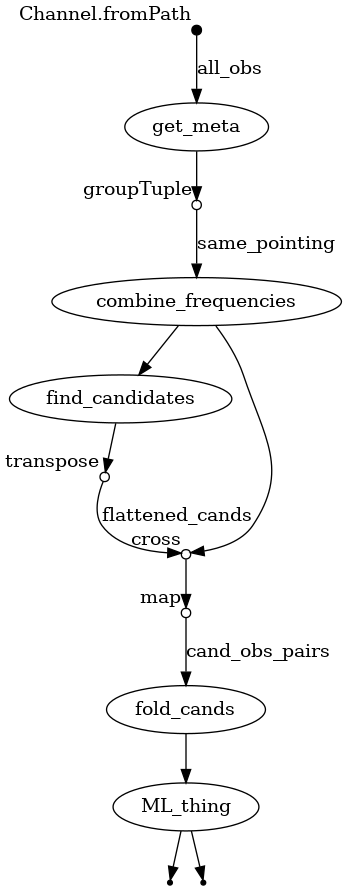 The graph isn’t super fancy, but you can see the processes as ellipses, the channels as linking lines, some with names, and circles on the channels where we have done some channel manipulation.
The graph isn’t super fancy, but you can see the processes as ellipses, the channels as linking lines, some with names, and circles on the channels where we have done some channel manipulation.
We can also review the report and timeline.
Key Points
get your channels right before working on the process
use
-resumewhile developing to reduce execution timereport and timeline can be used to tune the execution once your workflow actually works