Calculating Zonal Statistics on Rasters
Last updated on 2025-05-20 | Edit this page
Estimated time: 40 minutes
Overview
Questions
- How to compute raster statistics on different zones delineated by vector data?
Objectives
- Extract zones from the vector dataset
- Convert vector data to raster
- Calculate raster statistics over zones
Introduction
Statistics on predefined zones of the raster data are commonly used for analysis and to better understand the data. These zones are often provided within a single vector dataset, identified by certain vector attributes. For example, in the previous episodes, we defined infrastructure regions and built-up regions on Rhodes Island as polygons. Each region can be respectively identified as a “zone”, resulting in two zones. One can evualuate the effect of the wild fire on the two zones by calculating the zonal statistics.
Data loading
We have created assets.gpkg in Episode “Vector data in
Python”, which contains the infrastructure regions and built-up regions
. We also calculated the burned index in Episode “Raster Calculations in
Python” and saved it in burned.tif. Lets load them:
Align datasets
Before we continue, let’s check if the two datasets are in the same CRS:
OUTPUT
EPSG:4326
EPSG:32635The two datasets are in different CRS. Let’s reproject the assets to the same CRS as the burned index raster:
Rasterize the vector data
One way to define the zones, is to create a grid space with the same
extent and resolution as the burned index raster, and with the numerical
values in the grid representing the type of infrastructure, i.e., the
zones. This can be done by rasterize the vector data assets
to the grid space of burned.
Let’s first take two elements from assets, the geometry
column, and the code of the region.
OUTPUT
[[<POLYGON ((602761.27 4013139.375, 602764.522 4013072.287, 602771.476 4012998...>,
1],
[<POLYGON ((602779.808 4013298.838, 602772.497 4013266.01, 602768.577 4013242...>,
1],
[<POLYGON ((602594.855 4012962.661, 602593.423 4012983.028, 602588.485 401302...>,
1],
...]The raster image burned is a 3D image with a “band”
dimension.
OUTPUT
(1, 4523, 4828)To create the grid space, we only need the two spatial dimensions. We
can used .squeeze() to drop the band dimension:
OUTPUT
(4523, 4828)Now we can use features.rasterize from
rasterio to rasterize the vector data assets
to the grid space of burned:
PYTHON
from rasterio import features
assets_rasterized = features.rasterize(geom, out_shape=burned_squeeze.shape, transform=burned.rio.transform())
assets_rasterizedOUTPUT
array([[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
...,
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0]], dtype=uint8)Perform zonal statistics
The rasterized zones assets_rasterized is a
numpy array. The Python package xrspatial,
which is the one we will use for zoning statistics, accepts
xarray.DataArray. We need to first convert
assets_rasterized. We can use burned_squeeze
as a template:
PYTHON
assets_rasterized_xarr = burned_squeeze.copy()
assets_rasterized_xarr.data = assets_rasterized
assets_rasterized_xarr.plot()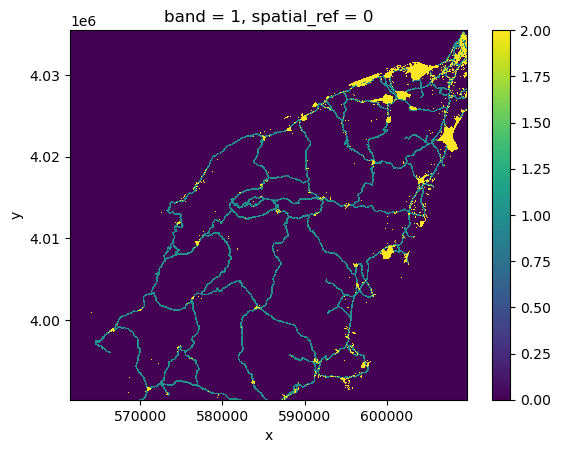
Then we can calculate the zonal statistics using the
zonal_stats function:
PYTHON
from xrspatial import zonal_stats
stats = zonal_stats(assets_rasterized_xarr, burned_squeeze)
statsOUTPUT
zone mean max min sum std var count
0 0 0.023504 1.0 0.0 485164.0 0.151498 0.022952 20641610.0
1 1 0.010361 1.0 0.0 9634.0 0.101259 0.010253 929853.0
2 2 0.000004 1.0 0.0 1.0 0.001940 0.000004 265581.0The results provide statistics for three zones: 1
represents infrastructure regions, 2 represents built-up
regions, and 0 represents the rest of the area.
- Zones can be extracted by attribute columns of a vector dataset
- Zones can be rasterized using
rasterio.features.rasterize - Calculate zonal statistics with
xrspatial.zonal_statsover the rasterized zones.
