Better research by better sharing
Figure 1
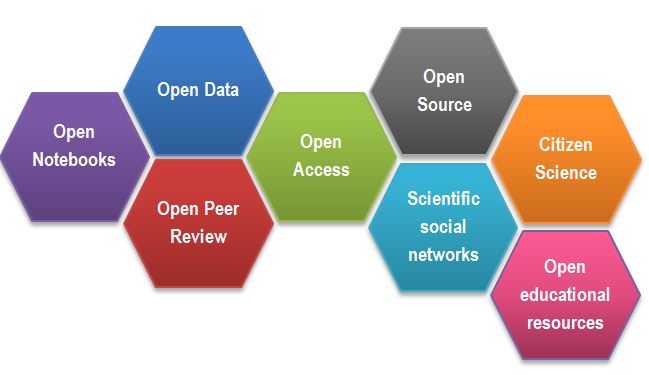
Figure 2
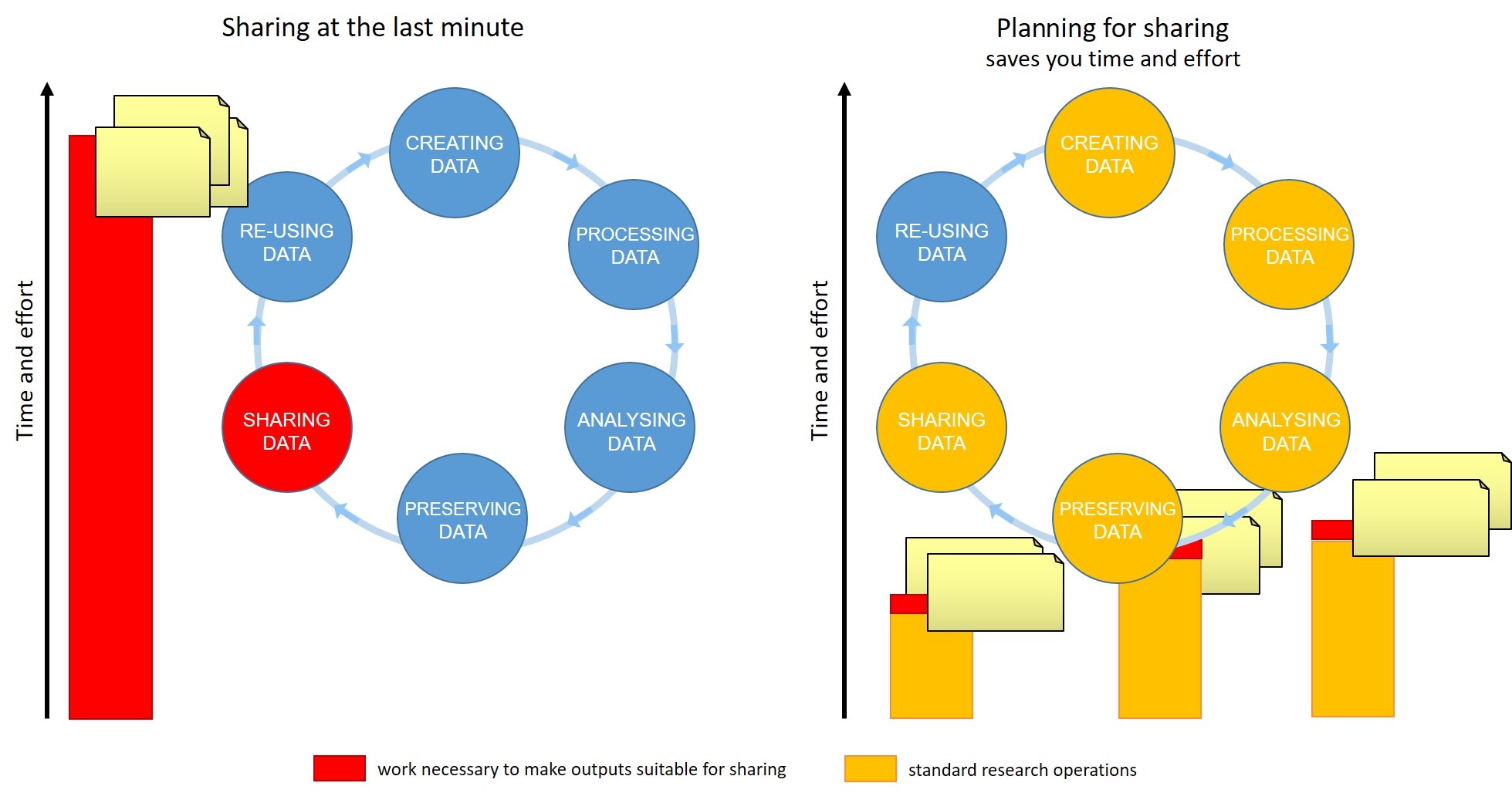 Figure credits:
Tomasz Zielinski and Andrés Romanowski
Figure credits:
Tomasz Zielinski and Andrés Romanowski
Being FAIR
Figure 1
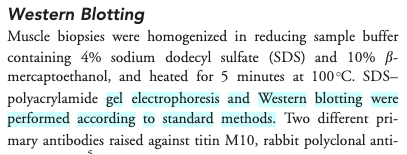
Figure 2
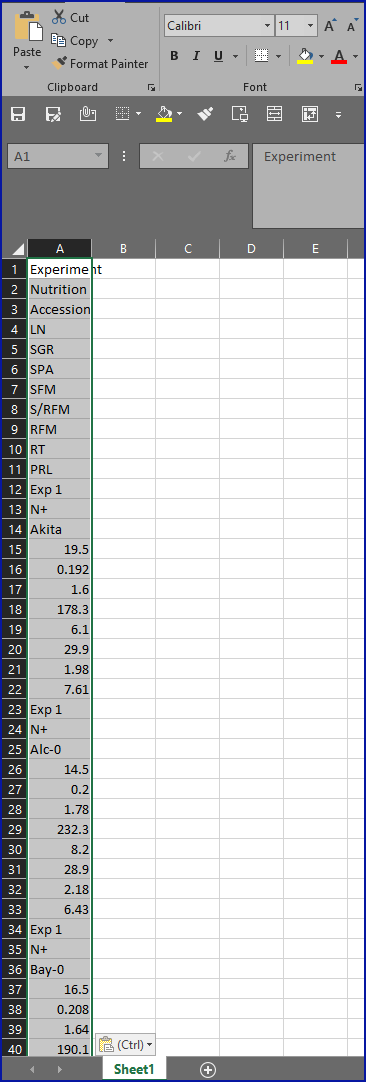
Figure 3
Puzzlingly, the authors have chosen to include the SRA accession
number in the final sentence of the ‘Acknowledgments’ section.
Perplexingly, searching the SRA website with the accession number brings
back twelve results at the time of writing, none of which contain the
cited accession number in them, but they appear to correspond to an
experiment with a title and institutional affiliation which match the
journal article, as you can see from this screenshot: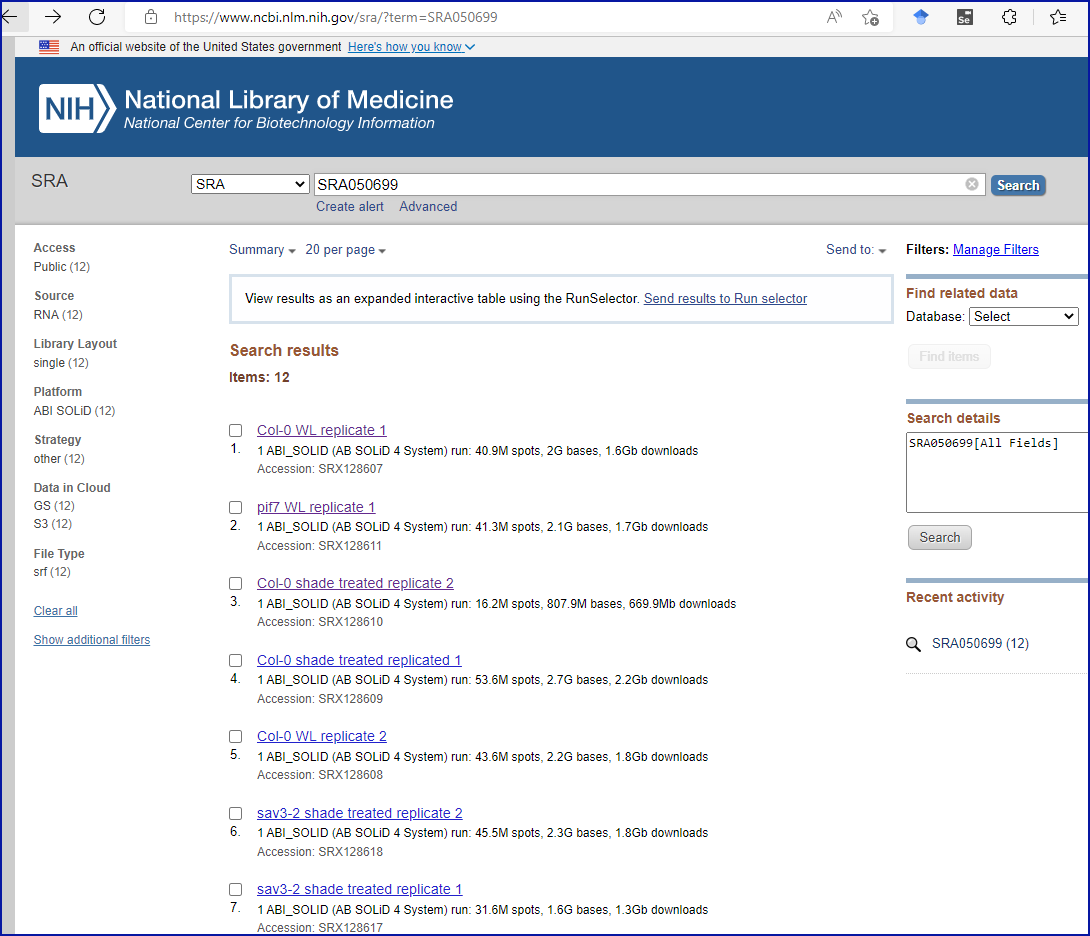
Figure 4
 From SangyaPundir
From SangyaPundir
Tools for Oracles and Overlords
Figure 1
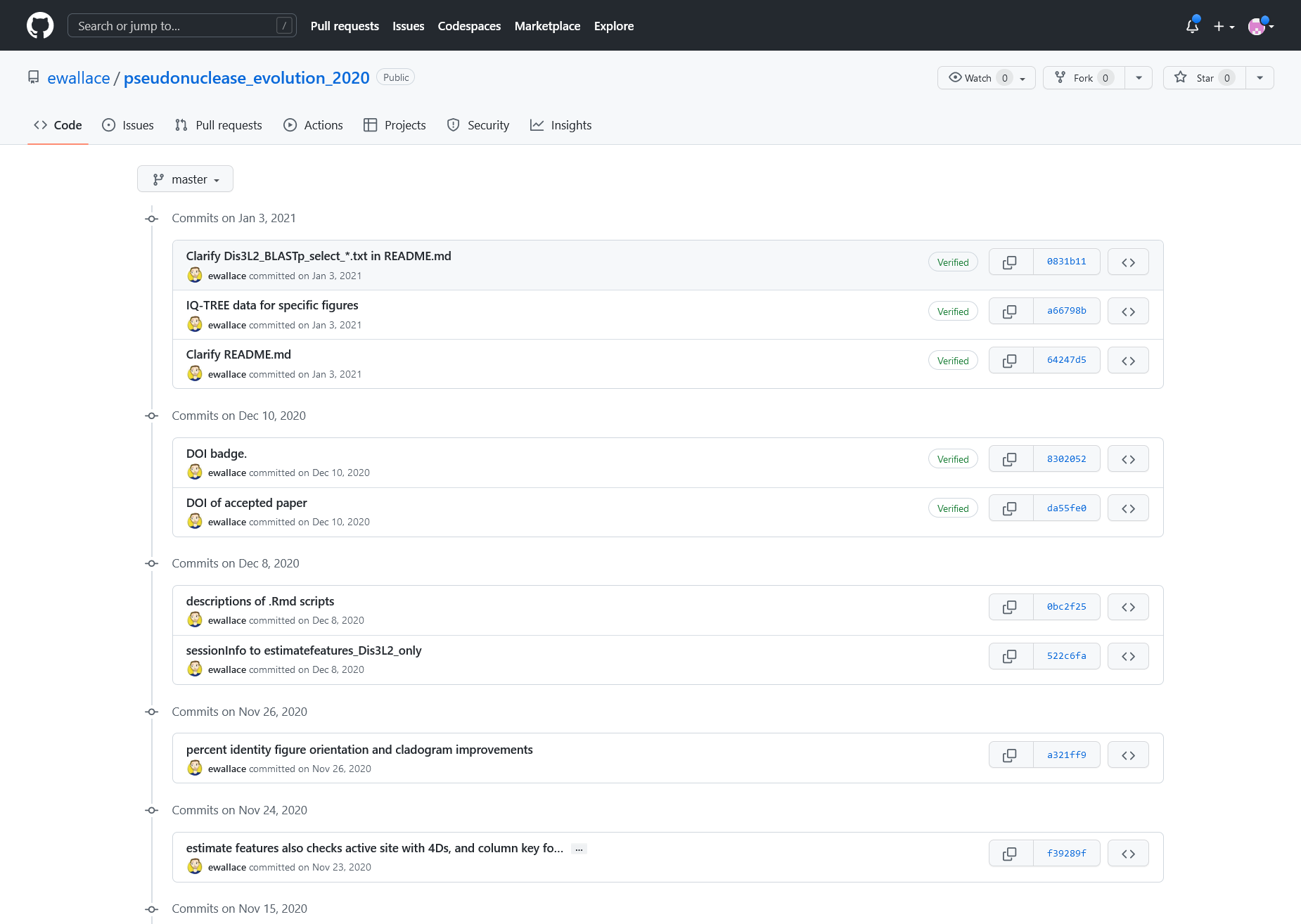
Figure 2
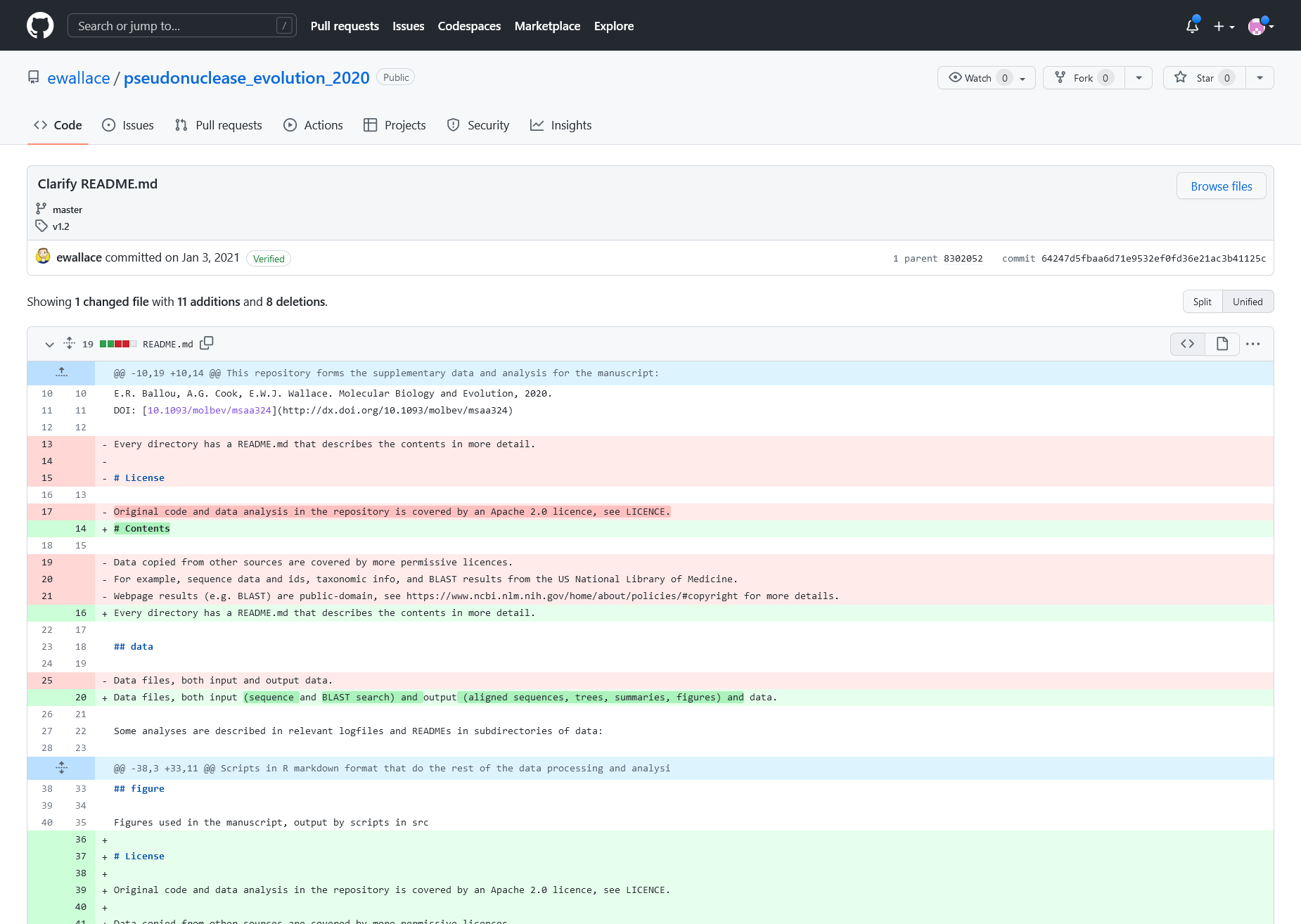
Figure 3
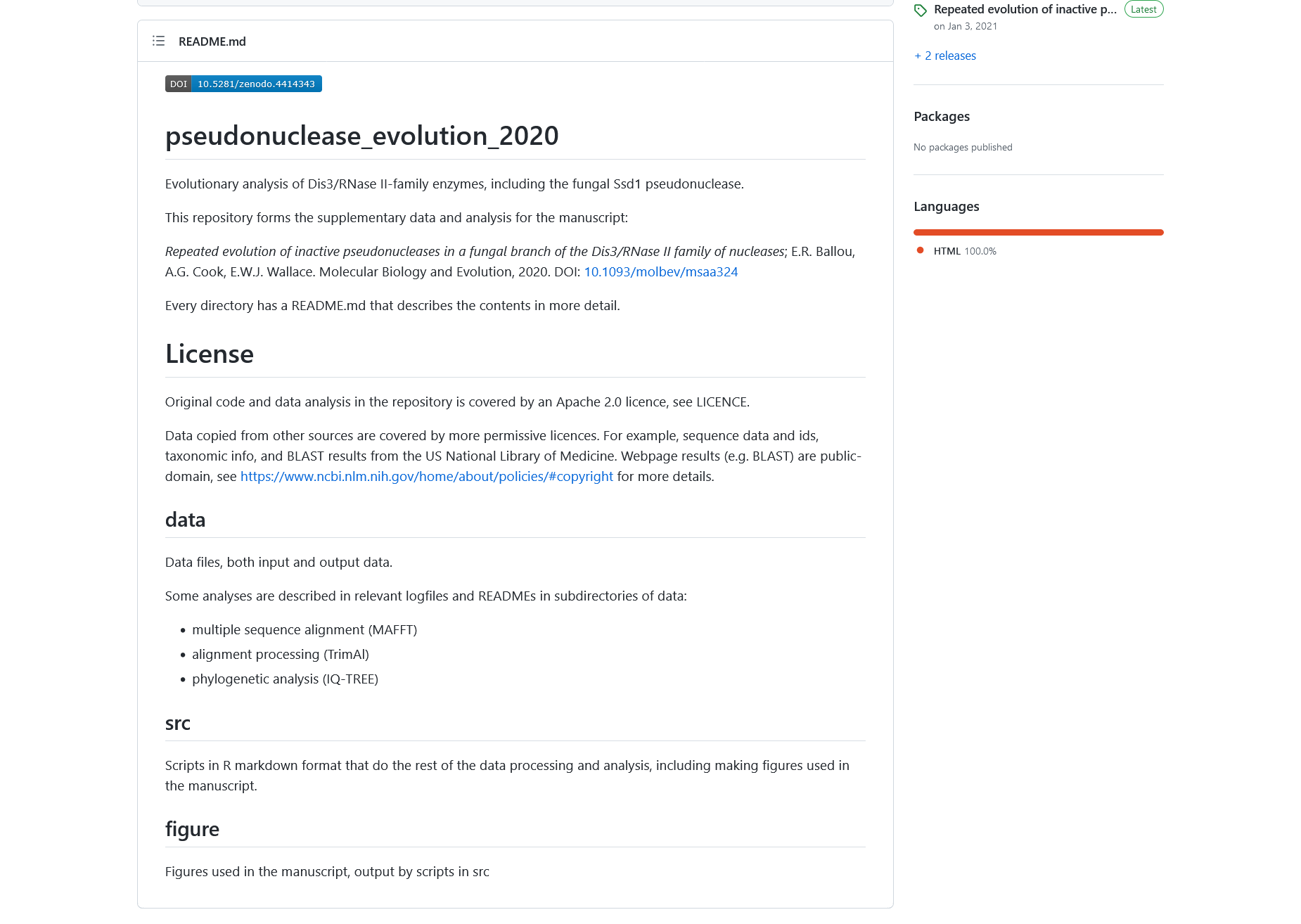
Figure 4
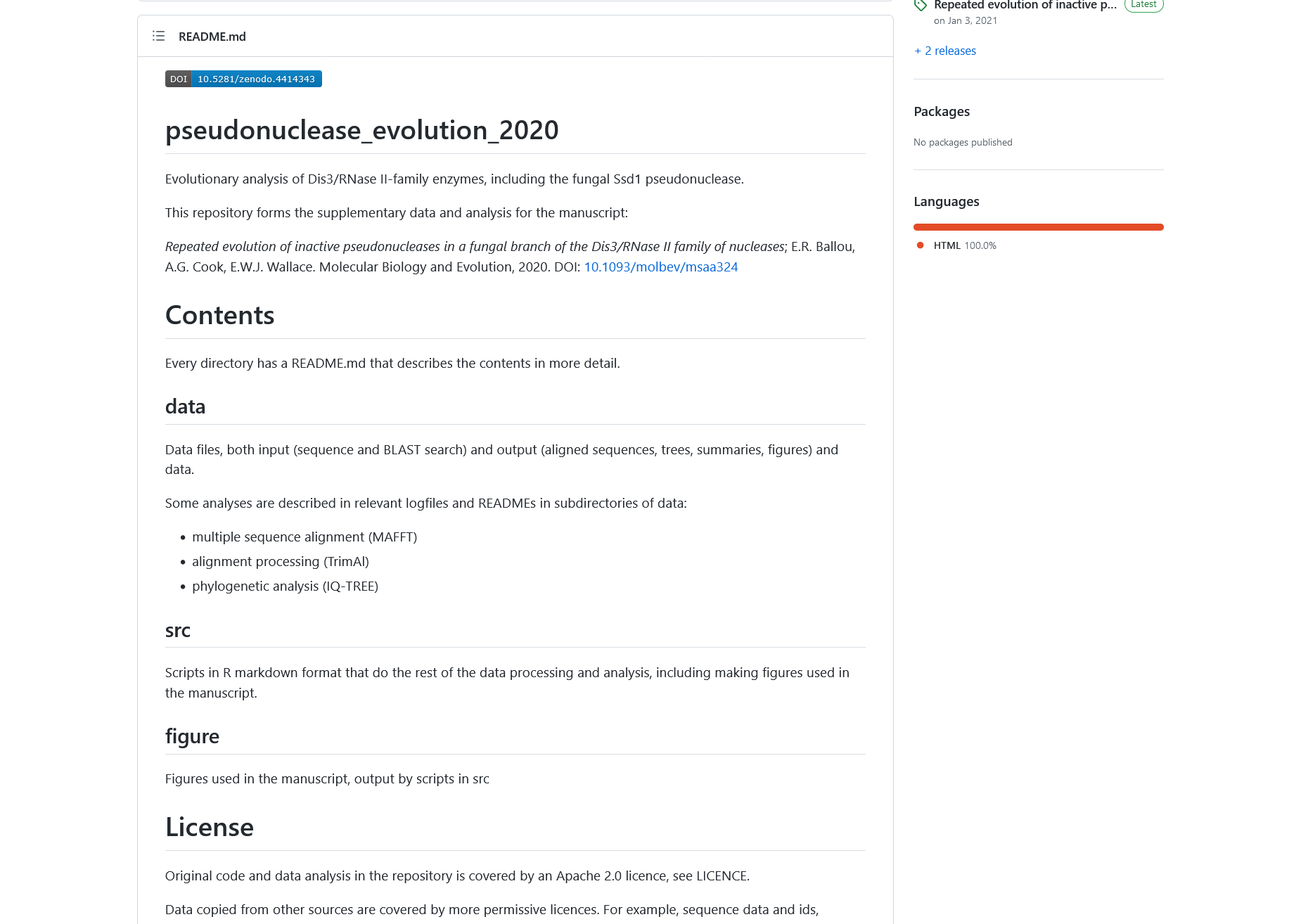
Figure 5
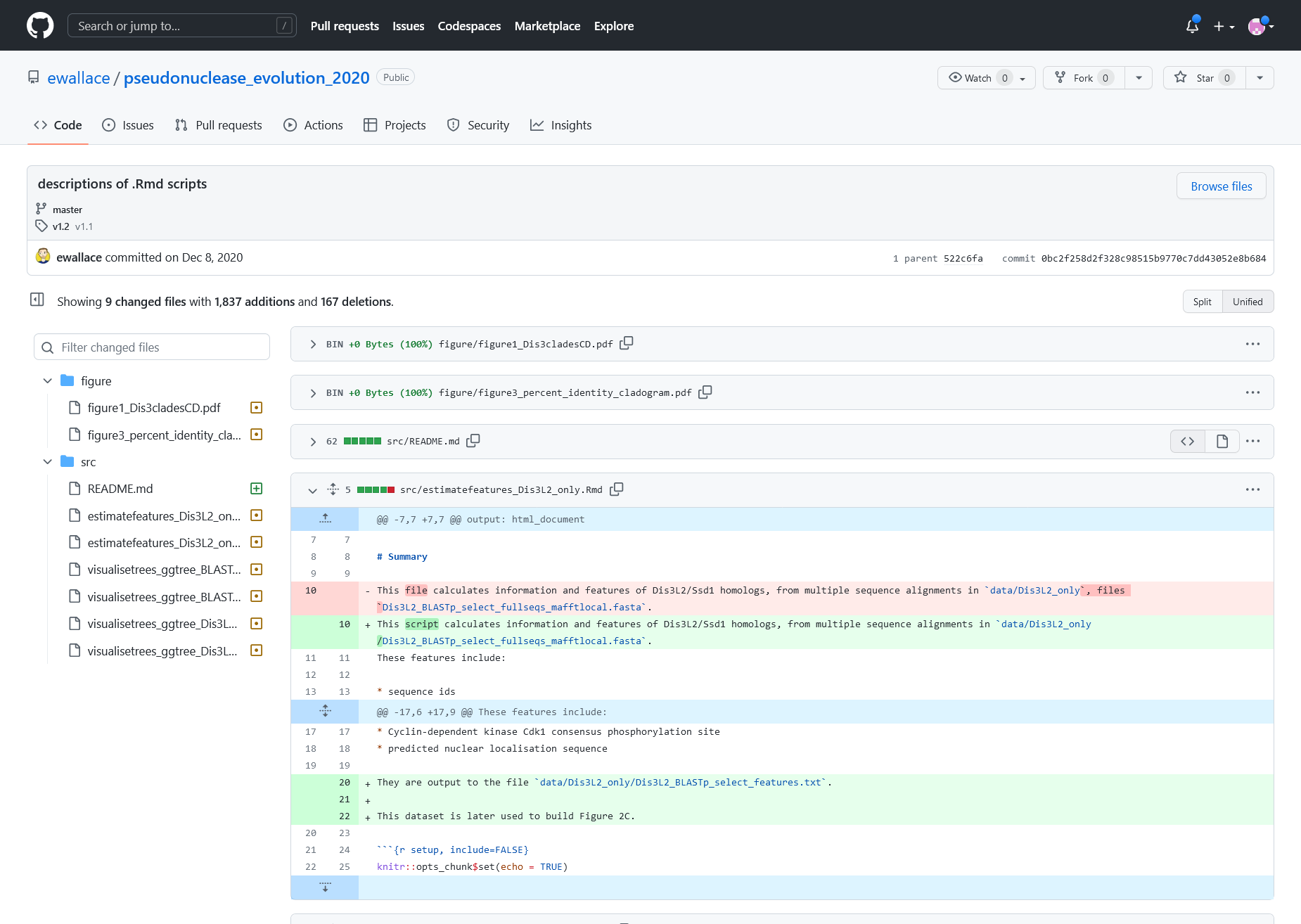
Figure 6
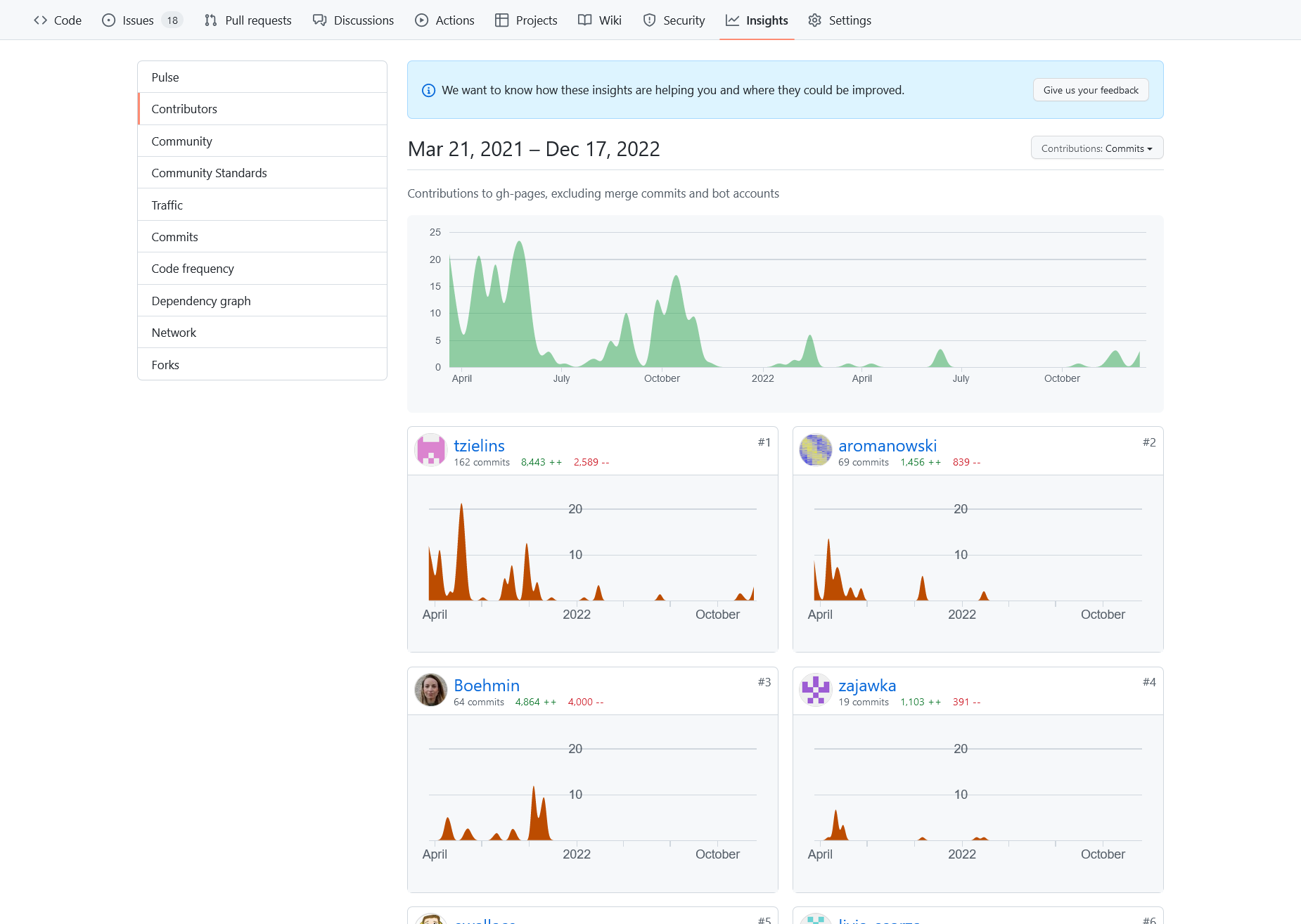
Figure 7
Figure 8
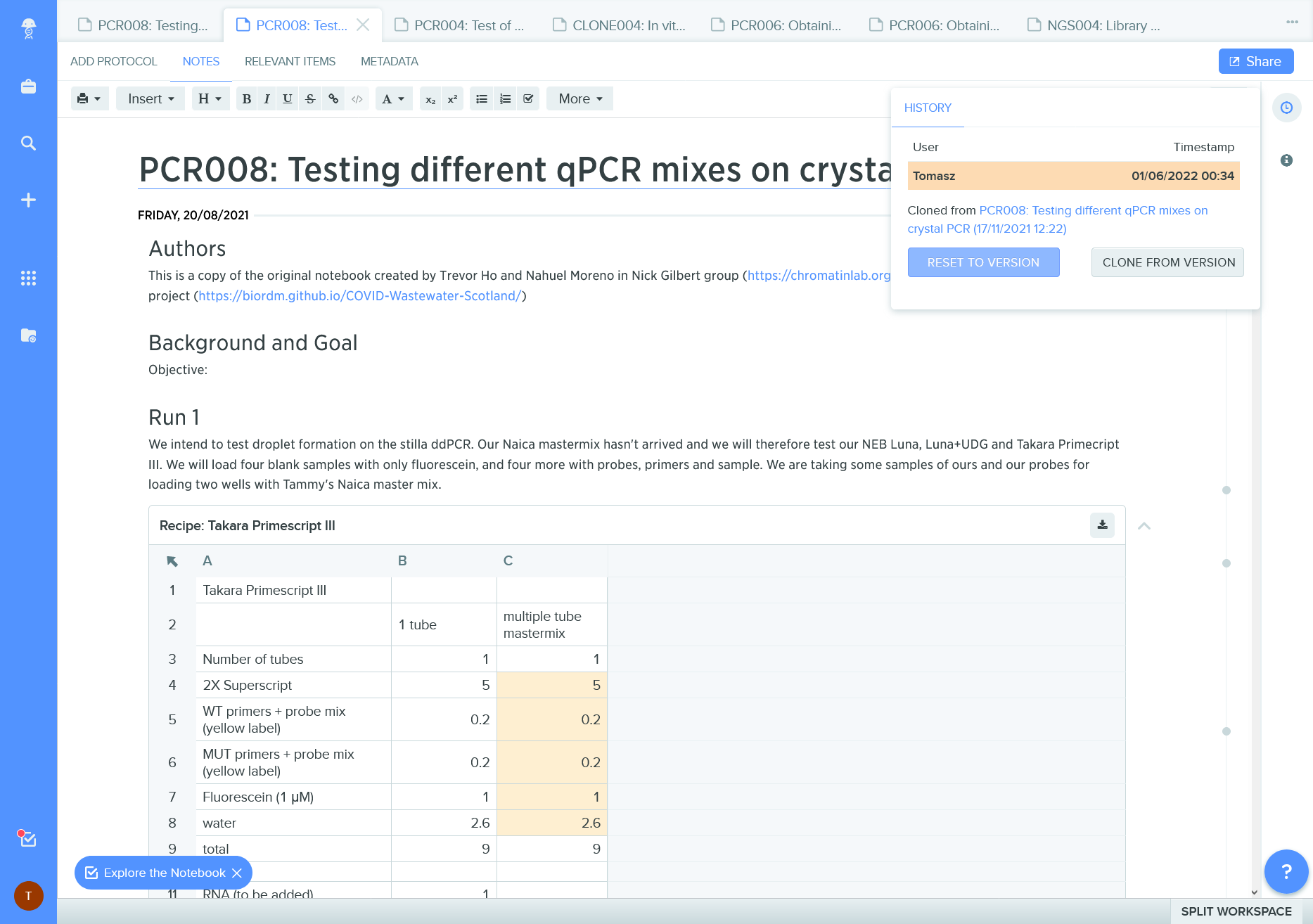
Figure 9
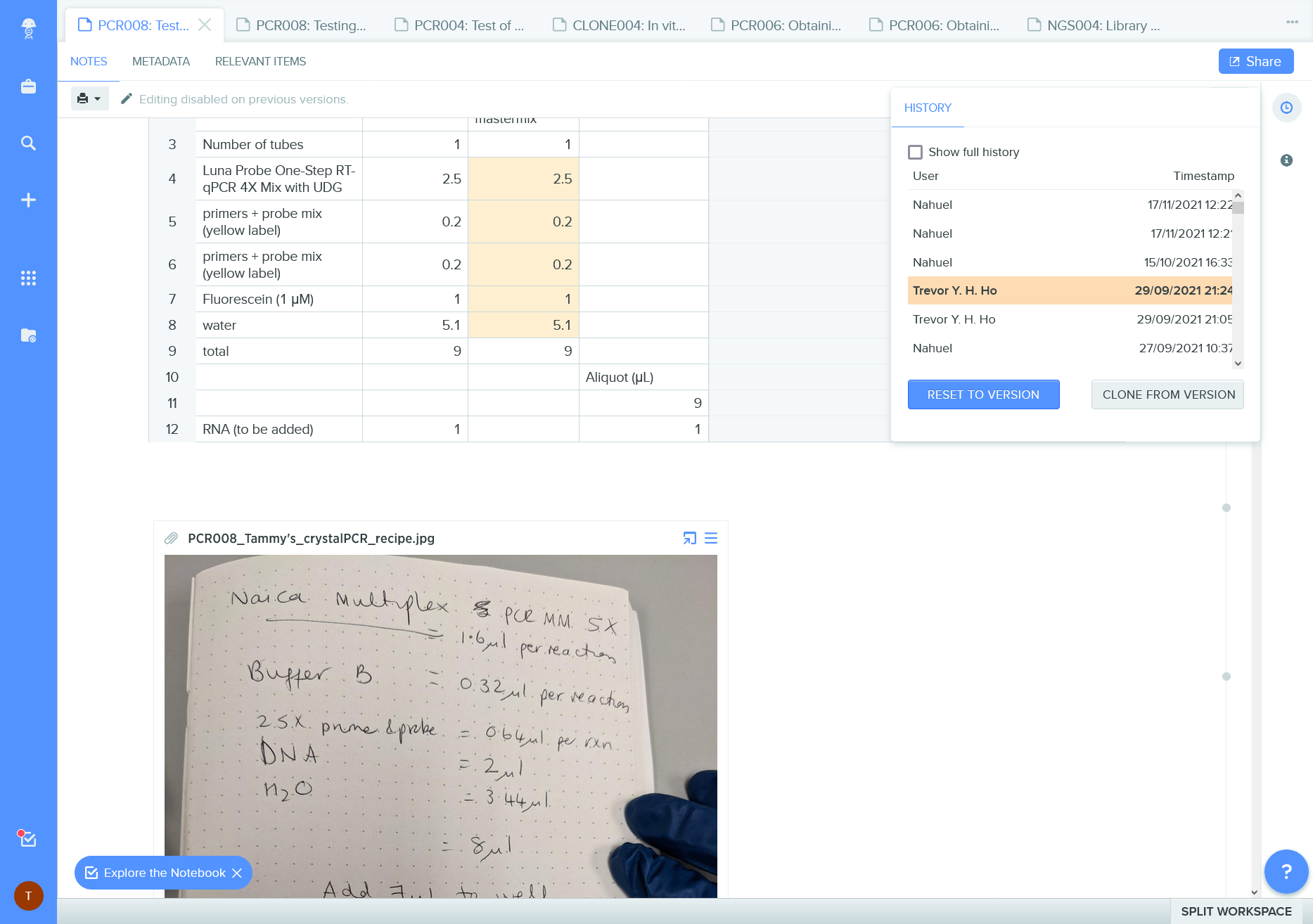
Figure 10
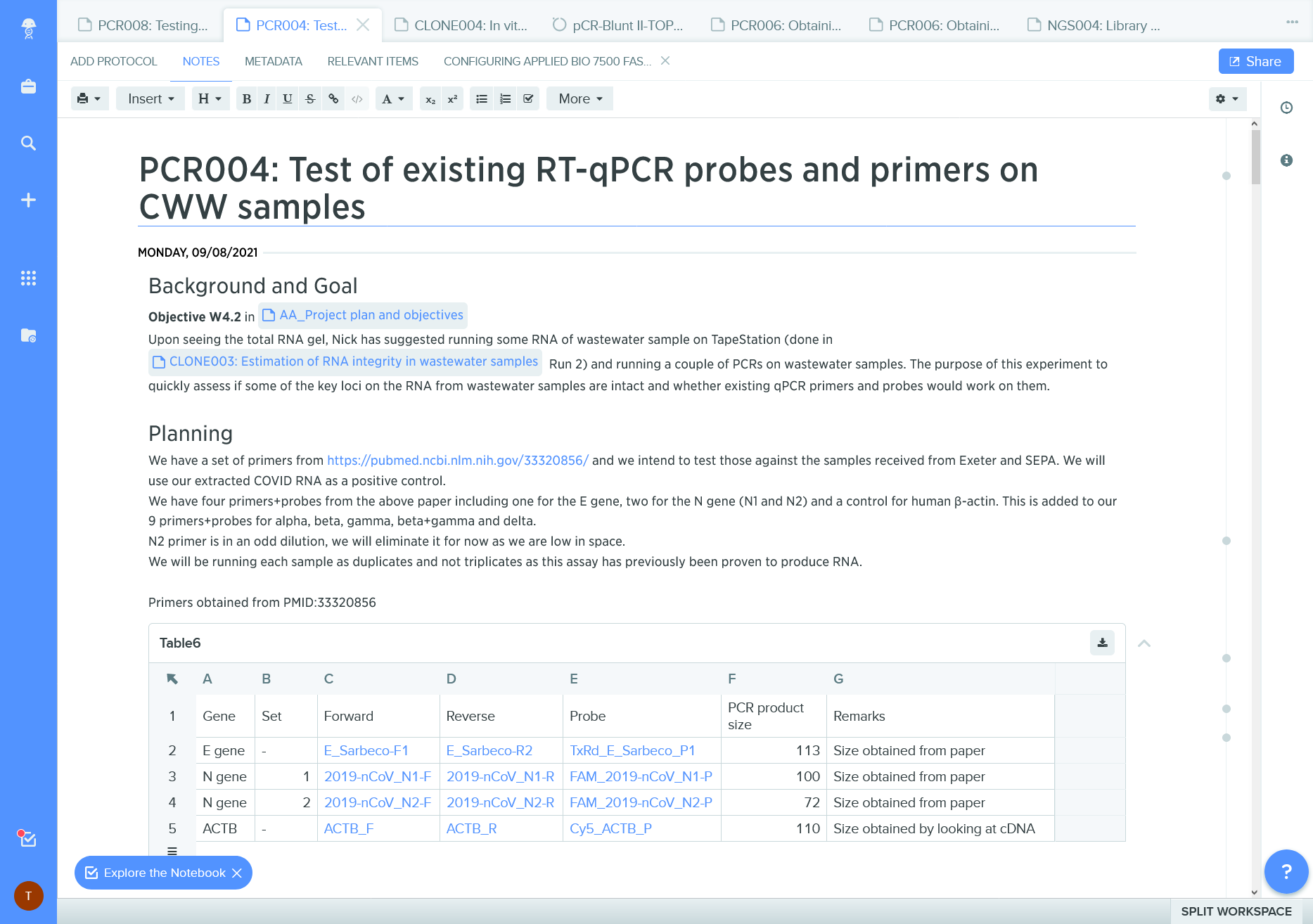
Figure 11
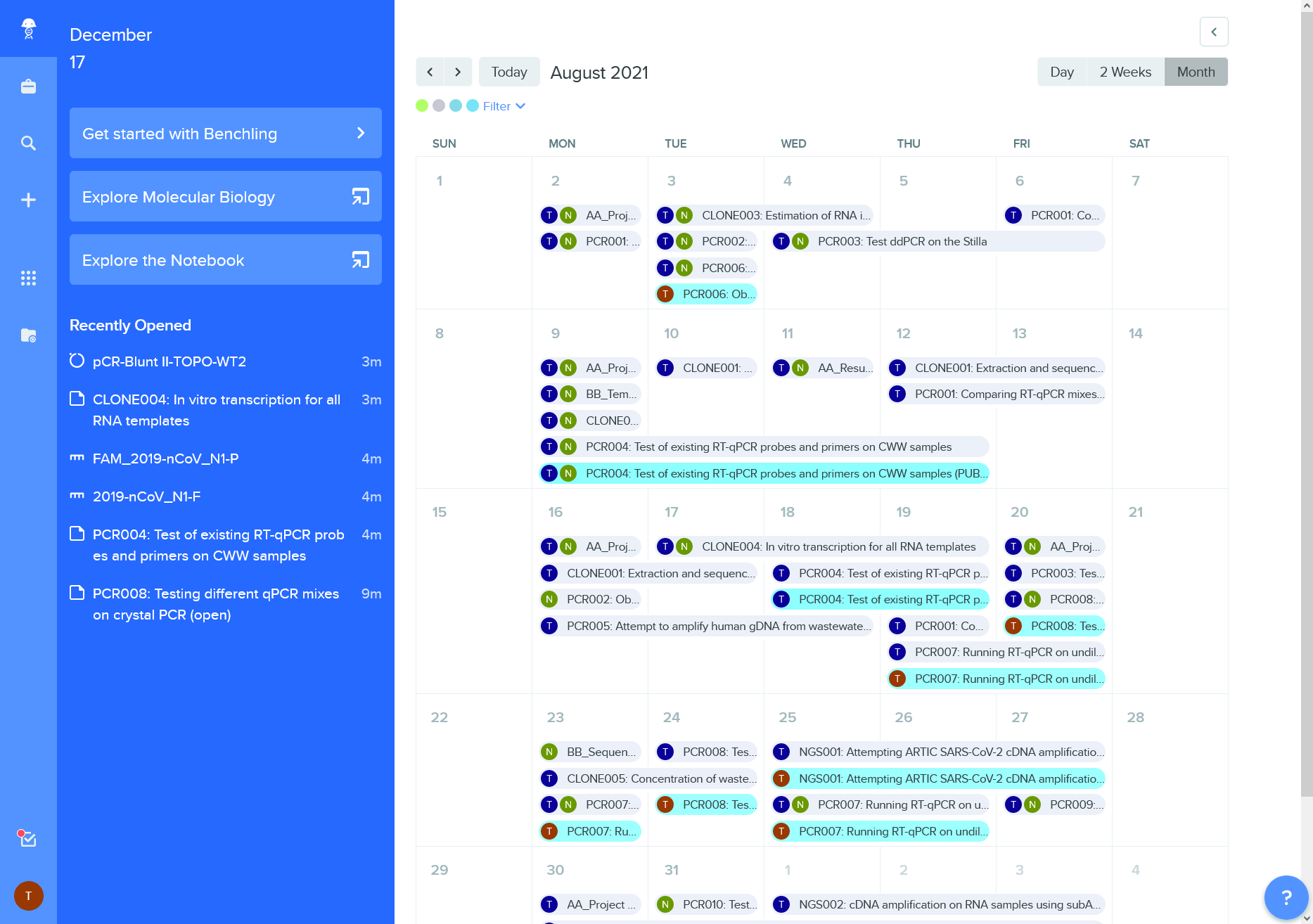
Figure 12
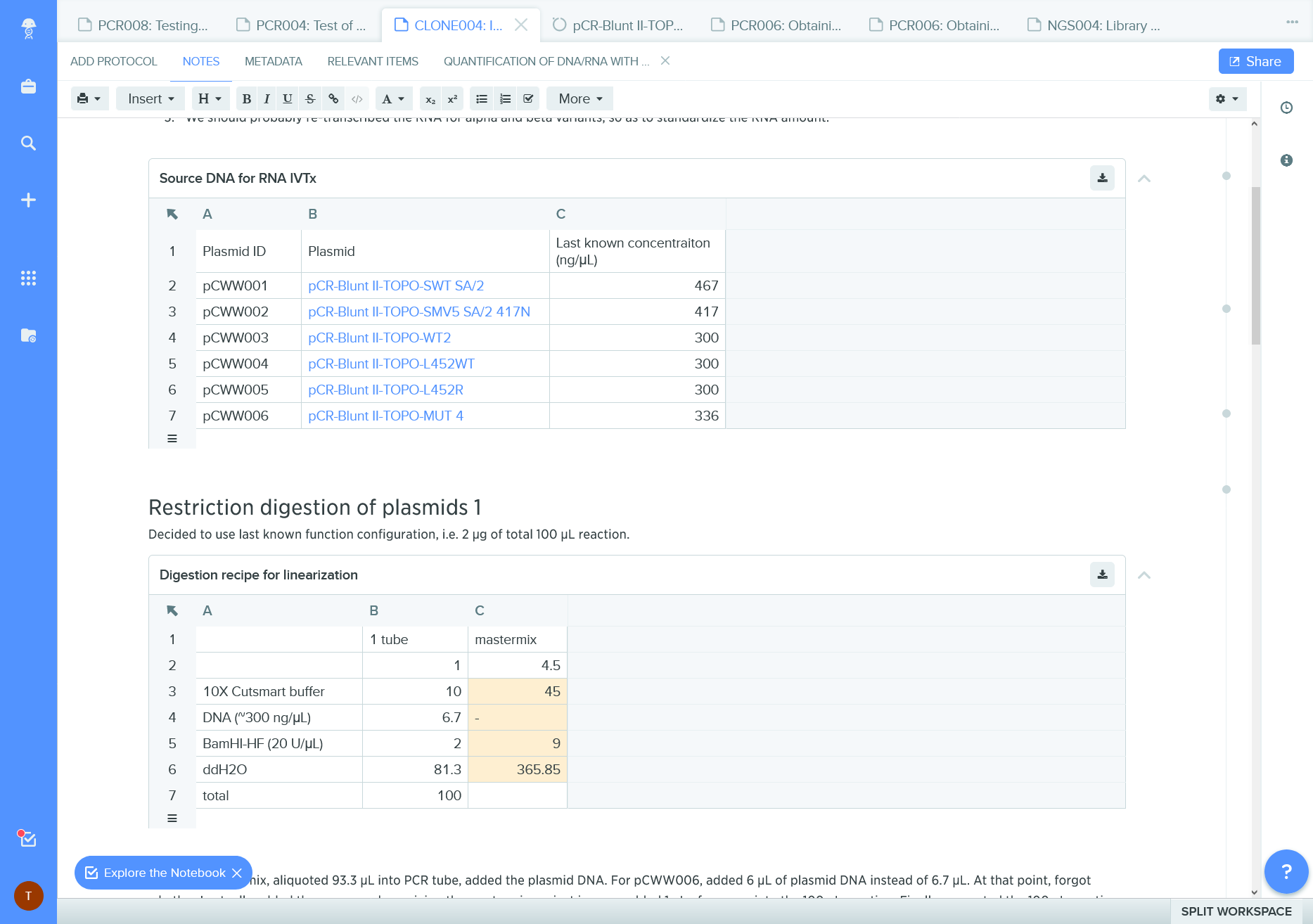
Figure 13
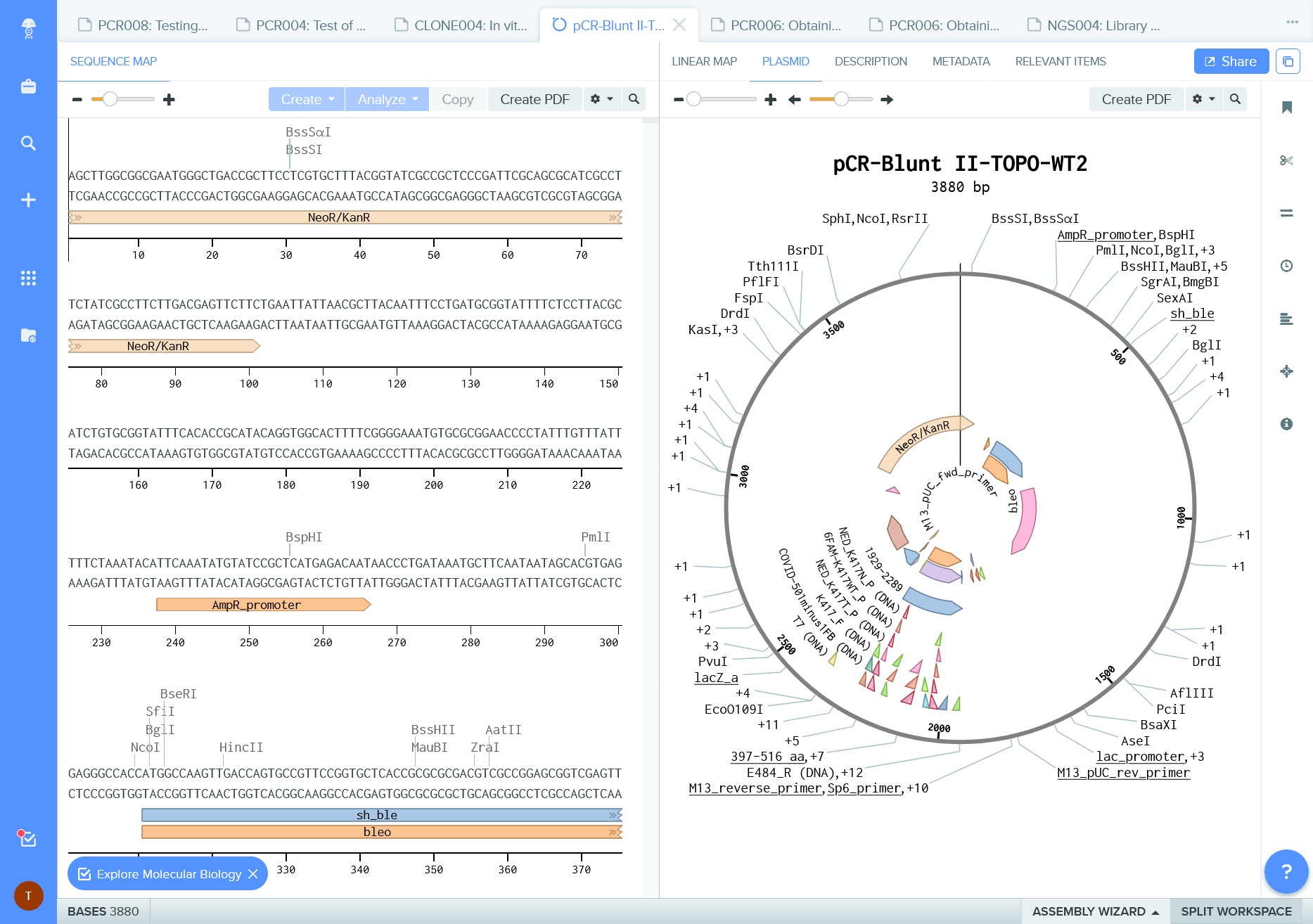
Public repositories
Figure 1
Figure 2
Figure 3
Taking another research area: if you search for ‘genomics’ on
FAIRsharing, you’ll get a list of more than fifty recommended
repositories.
Whereas PLoS provides a more biologically meaningful set of suggested
Omics repositories, see screenshot below: 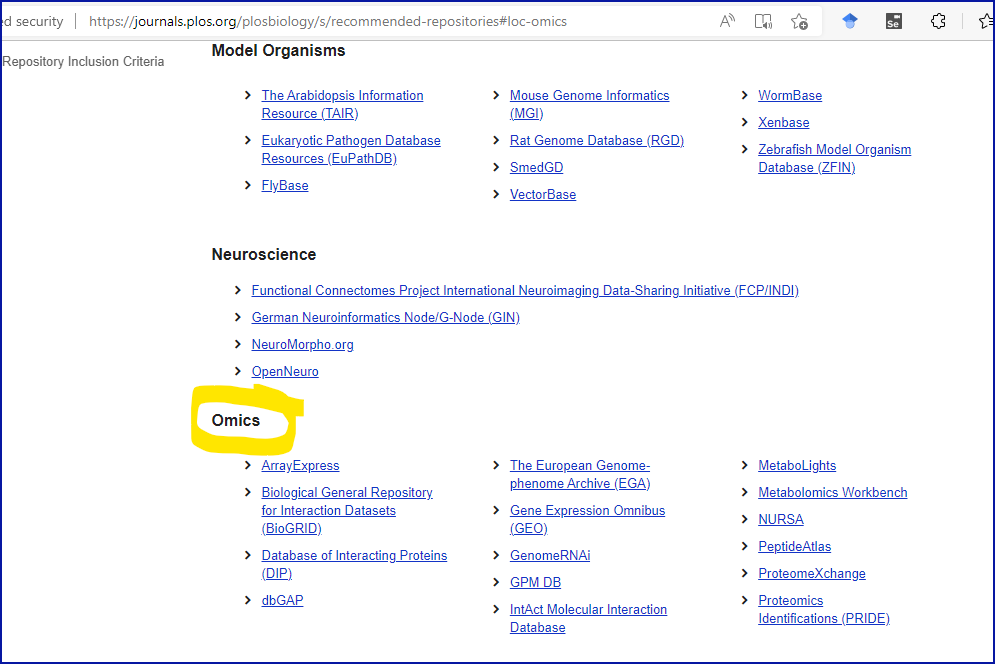
Template
Figure 1

