Chaining rules
Last updated on 2024-06-04 | Edit this page
Overview
Questions
- How do I combine steps into a workflow?
- How do I make a step that uses the outputs from a previous step?
Objectives
- Create workflow pipelines with dependencies.
- Use the outputs of one step as inputs to the next.
A pipeline of multiple rules
In this episode, we will plot the scaling results generated in the
last episode using the script plot_terse_amdahl_results.py.
These results report how long the work of running amdahl
took when using between 2 and 36 processes.
We want to plot these results automatically, i.e. as part of our workflow. In order to do this correctly, we need to make sure that our python plotting script runs only after we have finished calculating all results.
We can control the order and relative timing of when steps execute by defining dependencies.
For example, consider the following YAML,
depends.yaml:
YML
description:
name: Dependency-exploration
description: Experiment with adding dependencies
batch:
type: slurm
host: ruby # machine
bank: guests # bank
queue: pbatch # partition
reservation: HPCC1B # reservation for this workshop
env:
variables:
OUTPUT_PATH: ./Episode5
OUTPUT: date.txt
study:
- name: date-login
description: Write the date and login node's hostname to a file
run:
cmd: |
echo "From login node:" >> $(OUTPUT)
hostname >> $(OUTPUT)
date >> $(OUTPUT)
sleep 10
- name: date-batch
description: Write the date and node's hostname to a file
run:
cmd: |
echo "From batch node:" >> $(OUTPUT)
hostname >> $(OUTPUT)
date >> $(OUTPUT)
sleep 10
nodes: 1
procs: 1
walltime: "00:00:30"This script has two steps, each of which writes the type of node of
its host, the particular hostname, and the date/time before waiting for
10 seconds. The step date-login is run on a login node and
the step date-batch is run on a “batch” or “compute”
node.
Challenge
Run the above script with
You can then cat the output of both steps to
stdout via a command of the form
where {FILL} should be replaced by the date/time info of
this run. This will print the output of the two steps to
stdout.
Which step is printed first and how close together are they completed?
You’ll likely see output somewhat similar to
OUTPUT
From batch node:
pascal16
Tue Mar 26 14:36:04 PDT 2024
From login node:
pascal83
Tue Mar 26 14:36:03 PDT 2024though the times/dates and hostnames will be different. We didn’t try to control the order of operations, so each step will have run as soon as it had resources. Probably you’ll see that the timestamp on the login node is earlier because it will not have to wait for resources from the queue.
Next, let’s add a dependency to ensure that the batch step runs
before the login node step. To add a dependency a line
with the following format must be added to a step’s run
block:
{STEP NAME} is replaced by the name of the step from the
study that you want the current step to depend upon.
If we update date-login to include a dependency, we’ll
see
YML
- name: date-login
description: Write the date and login node's hostname to a file
run:
cmd: |
echo "From login node:" >> $(OUTPUT)
hostname >> $(OUTPUT)
date >> $(OUTPUT)
sleep 10
depends: [date-batch]Now date-login will not run until
date-batch has finished.
Challenge
Update depends.yaml to make date-login wait
for date-batch to complete before running. Then rerun
maestro run depends.yaml.
How has the output of the two date.txt files
changed?
This time, you should see that the date printed from the login node is at least 10 seconds later than the date printed on the batch node. For example, on Pascal I see
OUTPUT
From batch node:
pascal16
Tue Mar 26 15:09:54 PDT 2024
From login node:
pascal83
Tue Mar 26 15:10:53 PDT 2024Callout
Slurm can also be used to define dependencies. How is using Maestro to define the order of our steps any different?
One difference is that we can use Maestro to order steps that
are not seen by Slurm. Above, date-login was run
on the login node. It wasn’t submitted to the queue, and Slurm never saw
that step. (It wasn’t given a Slurm job ID, for example.) Maestro can
control the order of steps running both in and outside the batch queue,
whereas Slurm can only enforce dependencies between Slurm-scheduled
jobs.
A step that waits for all iterations of its dependency
Let’s return to our Amdahl scaling study and the YAML with which we ended in the last episode:
YML
description:
name: Amdahl
description: Run a parallel program
batch:
type: slurm
host: ruby # machine
bank: guests # bank
queue: pbatch # partition
reservation: HPCC1B # reservation for this workshop
env:
variables:
P: .999
OUTPUT: amdahl.json
OUTPUT_PATH: ./Episode5
study:
- name: amdahl
description: run in parallel
run:
# Here's where we include our MPI wrapper:
cmd: |
$(LAUNCHER) amdahl --terse -p $(P) >> $(OUTPUT)
nodes: 1
procs: $(TASKS)
walltime: "00:00:30"
global.parameters:
TASKS:
values: [2, 4, 8, 18, 24, 36]
label: TASKS.%%Ultimately we want to add a plotting step that depends upon
amdahl, but for now let’s create a placeholder that will go
under study and beneath amdahl:
YML
- name: plot
description: Create a plot from `amdahl` results
run:
# We'll update this `cmd` later
cmd: |
echo "This is where we plot"Based on what we saw before, we might think that we just need to add
to the end of this block. Let’s try this to see what happens.
Doing a dry run with amdahl.yaml (text below) should
generate an Amdahl_{Date & time stamp} directory with a
subdirectory for the plot step. Within the ‘plot’
subdirectory, there will be several TASKS.%% subdirectories
– one for each of the values of TASKS defined under
global.parameters in amdahl.yaml. For
example,
OUTPUT
~/Episode5/Amdahl_20240429-153515OUTPUT
amdahl Amdahl.study.pkl batch.info meta plot
Amdahl.pkl Amdahl.txt logs pascal-amdahl.yaml status.csvOUTPUT
TASKS.18 TASKS.2 TASKS.24 TASKS.36 TASKS.4 TASKS.8OUTPUT
TASKS.18/plot_TASKS.18.slurm.shEach of these TASKS.%% subdirectories – such as
plot/TASKS.18 – represents a separate workflow step that is
related to its corresponding amdahl/TASKS.%% workflow step
and directory.
The text of the yaml used is below.
YML
description:
name: Amdahl
description: Run a parallel program
batch:
type: slurm
host: ruby # machine
bank: guests # bank
queue: pbatch # partition
reservation: HPCC1B # reservation for this workshop
env:
variables:
P: .999
OUTPUT: amdahl.json
OUTPUT_PATH: ./Episode5
study:
- name: amdahl
description: run in parallel
run:
# Here's where we include our MPI wrapper:
cmd: |
$(LAUNCHER) amdahl --terse -p $(P) >> $(OUTPUT)
nodes: 1
procs: $(TASKS)
walltime: "00:00:30"
- name: plot
description: Create a plot from `amdahl` results
run:
# We'll update this `cmd` later
cmd: |
echo "This is where we plot"
depends: [amdahl]
global.parameters:
TASKS:
values: [2, 4, 8, 18, 24, 36]
label: TASKS.%%The takeaway from the above challenge is that, a step depending upon
a parameterized step will become parameterized by default. In this case,
creating a plotting step that depends on the amdahl step
will lead to a series of plots, each of which will use data only from a
single run of amdahl.
This is not what we want! Instead, we want to generate a plot that
uses data from several runs of amdahl – each of which will
use a different number of tasks. This means that plot
cannot run until all runs of amdahl have completed.
So, the syntax for defining dependency will change when parameterized
steps are involved. To indicate that we want plot to run
after ALL amdahl steps, we’ll add a
_* to the end of the step name:
Now our new step definition will look like
YML
- name: plot
description: Create a plot from `amdahl` results
run:
# We'll update this `cmd` later
cmd: |
echo "This is where we plot"
depends: [amdahl_*]Challenge
Update your amdahl.yaml so that plot runs
after amdahl has run with all values of TASKS.
Perform a dry run to verify that plot will run only
once.
Your new directory structure should look something like
OUTPUT
amdahl Amdahl.study.pkl batch.info meta plot
Amdahl.pkl Amdahl.txt logs pascal-amdahl.yaml status.csvOUTPUT
plot.slurm.shIn other words, the directory plot will have no
subdirectories.
Using the outputs from a previous step
Manually plotting scaling results
In your working directory, you should have a copy of
plot_terse_amdahl_results.py. The syntax for running this
script is
Before trying to add this command to our workflow, let’s run it
manually to see how it works. We can call the output image
output.jpg. As for the input names, we can use the
.json files created at the end of episode 4. In particular
if you run
using the <Date> and <Time> of
your last run in Episode 4, you should see a list of files named
amdahl.json:
OUTPUT
Episode4/Amdahl_20240326-155434/amdahl/TASKS.18/amdahl.json
Episode4/Amdahl_20240326-155434/amdahl/TASKS.24/amdahl.json
Episode4/Amdahl_20240326-155434/amdahl/TASKS.2/amdahl.json
Episode4/Amdahl_20240326-155434/amdahl/TASKS.36/amdahl.json
Episode4/Amdahl_20240326-155434/amdahl/TASKS.4/amdahl.json
Episode4/Amdahl_20240326-155434/amdahl/TASKS.8/amdahl.jsonYou can use this same filepath with wildcards to specify this list of JSON files as inputs to our python script:
BASH
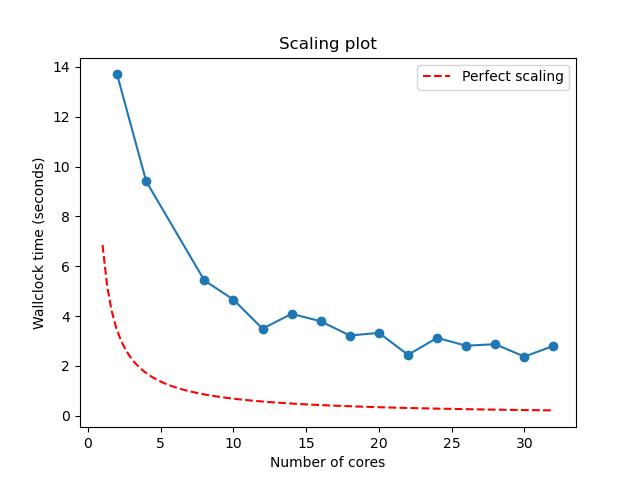
python3 plot_terse_amdahl_results.py output.jpg Episode4/Amdahl_<Date>_<Time>/amdahl/TASKS.*/amdahl.jsonChallenge
Generate a scaling plot by manually specifying the JSON files
produced from a previous run of amdahl.yaml.
The resulting JPEG should look something like
Adding plotting to our workflow
Let’s update our plot step in amdahl.yaml
to include python plotting rather than a placeholder echo
command. We want the updated step to look something like
YML
- name: plot
description: Create a plot from `amdahl` results
run:
cmd: |
python3 plot_terse_amdahl_results.py output.jpg Episode5/Amdahl_<Date>_<Time>/amdahl/TASKS.*/amdahl.json
depends: [amdahl_*]The trouble is that we don’t know the exact value of
Episode5/Amdahl_<Date>_<Time>/amdahl for a job
that we haven’t run yet. Luckily Maestro gives us a placeholder to the
equivalent of this path for the current job –
$(amdahl.workspace). This is the workspace for the
amdahl step, where all outputs for amdahl,
including our TASKS.* directories, are written.
This means we can update the plot step as follows:
YML
- name: plot
description: Create a plot from `amdahl` results
run:
cmd: |
python3 plot_terse_amdahl_results.py output.jpg $(amdahl.workspace)/TASKS.*/amdahl.json
depends: [amdahl_*]Callout
Where does the plot_terse_amdahl_results.py script live?
In Maestro, $(SPECROOT) specifies the root directory from
which you originally ran maestro run .... This is where
plot_terse_amdahl_results.py should live, so let’s be more
precise:
Challenge
Update amdahl.yaml so that
- one step definition runs
amdahlfor 50% parallelizable code using [2, 4, 8, 16, 32] tasks - a second step plots the results.
Your YAML file should look something like
YML
description:
name: Amdahl
description: Run a parallel program
batch:
type: slurm
host: ruby # machine
bank: guests # bank
queue: pbatch # partition
reservation: HPCC1B # reservation for this workshop
env:
variables:
P: .5
OUTPUT: amdahl.json
OUTPUT_PATH: ./Episode5
study:
- name: run-amdahl
description: run in parallel
run:
# Here's where we include our MPI wrapper:
cmd: |
$(LAUNCHER) amdahl --terse -p $(P) >> $(OUTPUT)
nodes: 1
procs: $(TASKS)
walltime: "00:01:30"
- name: plot
description: Create a plot from `amdahl` results
run:
cmd: |
python3 $(SPECROOT)/plot_terse_amdahl_results.py output.jpg $(run-amdahl.workspace)/TASKS.*/amdahl.json
depends: [amdahl_*]
global.parameters:
TASKS:
values: [2, 4, 8, 16, 32]
label: TASKS.%%Errors are normal
Don’t be disheartened if you see errors when first testing your new
Maestro pipelines. There is a lot that can go wrong when writing a new
workflow, and you’ll normally need several iterations to get things just
right. Luckily, Maestro will do some checks for consistency
at the outset of the run. If you specify a dependency that doesn’t exist
(because of a rename, for example), the job will fail before submitting
work to the queue.
Key Points
- We can control the order of steps run in a study by creating dependencies.
- You can create a dependency with the
depends: [{step name}]syntax. - Dependency syntax changes to
depends: [{step name}_*]for parameterized steps.
