Data Sharing
Last updated on 2024-11-19 | Edit this page
Estimated time: 12 minutes
Overview
Questions
- What data repository would you publish your data in?
- What are the benefits of sharing you data?
- Do you need to include all your data in a repository?
Objectives
- Understand the benefits of sharing your research data and how to do that.
- Understand the different type’s of repositories available.
- Determine how to limit your dataset to be more usable.
Introduction
Sharing data that underlies research has become a common expectation within scholarly research. However, the landscape of data repositories is still uneven and many researchers are still learning best practices for data sharing.
Pick a Data Repository
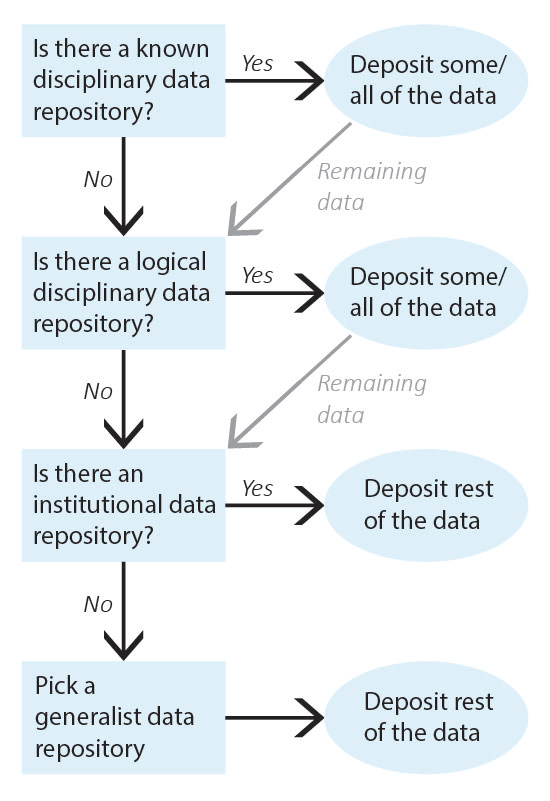
It can be difficult to know where to share research data as so many sharing platforms are available. Current guidance is to deposit data in data repository that will give you a DOI or similar permanent identifier. For this purpose, the University of Canterbury offers Figshare as our recommended general repository. The following exercise guides you through the process of picking a data repository, starting with repositories for very specific types of data and defaulting to generalist data repositories.
Repository fees
Some repositories charge fees for deposit, most often for large data (500 GB or larger). Figshare is free to UC Researchers, but currently has a limit of 1 TB of data. For more information see the UC figshare libguides.
Pick a Data Repository
Identify the data that needs to be shared and work through repository selection from discipline-specific data repositories to more general data repositories. Once you have identified a repository for all of your data, deposit the data and record the corresponding permanent identifiers. Note that, depending on data types, you may need to deposit your data into multiple repositories (for example, a discipline-specific repository for one type of data and an institutional data repository for the rest of the data).
1. Identify all of the data that needs to be shared.
Example: My data to be shared includes: 1) genetic data for Drosophila; and 2) microscope images of flies.
2. Is there a known disciplinary data repository for some or all of the data? For example, is there a data repository used by everyone in your research area or required for your data type by your funding agency?
If so, deposit some or all of your data there. Go to step 7 if the repository will accept all of your data or go to the next question if there is still some data left to deposit.
Example: The database FlyBase is used for Drosophila genes and genomes. My genetic data will be shared there.
3. Review the list of recommended data repositories from PLOS1. Is there a logical disciplinary data repository for some or all of your data?
If so, deposit some or all of your data there. Go to step 7 if you have shared all of your data or go to the next question if there is still some data left to deposit.
Example: There isn’t a logical disciplinary data repository for microscope images of flies.
4. Does your institution have a data repository?
If so, deposit the remainder of your data there and go to step 7.
Example: University of Canterbury Figshare. I will deposit my microscope images in UC Figshare.
5. Do you have a preferred generalist data repository2?
If so, deposit the remainder of your data there and go to step 7.
Example: [All data has been shared already.]
6. Pick a generalist data repository3 and deposit the remainder of your data.
Deposit your data and go to step 7.
Example: [All data has been shared already.]
7. Record the permanent identifier, ideally a DOI, from each data deposit.
DOIs make data FAIR4 and aid with data sharing compliance. If you did not receive a permanent identifier (such as a DOI, permanent URL, etc.) during deposit, return to step 2 and pick a different data repository for your data.
Example: UC Figshare provides DOIs for all deposits; my permanent identifier is doi.org/10.22002/XXXXXXXXXXX.
Share Data
Data sharing is becoming common and expected by funding agencies and journals. While the process of depositing data into a data repository will vary between repositories, there are some common actions that should be taken to prepare data for sharing.
What data to include in a dataset
While sharing data for a paper in a public repository is good (and sometimes required), there is little guidance on what data should be uploaded. Data to include in a repository is field specific, but we will present some guidelines here following the policies of the Nature Portfolio journals6.
Authors should provide their data at a level ‘rawness’ that allows it to be re-used, in alignment with accepted norms within their community. It may be advantageous to release some types of data at multiple levels to enable wider reuse – for example, proteomics data may best be released as ‘raw’ spectra as well as more processed peptide- or protein-level data – however this is not mandated as long as the level of ‘rawness’ is sufficient for some potential use. Authors may submit supplementary information files – including code (also see our code availability policy), models, workflows and summary tables – however we strongly encourage deposition in repositories as a first preference, especially for primary data, which should not be submitted as supplementary information.
In short, you should upload data to the repository in the rawest format that allows for a reader to reproduce your results. This means the data shouldn’t be filtered or have any sort of operations done to it. To illustrate what this might look like, let’s go over a real-life case study.
Data Case Study - Computational Fluid Dynamics Simulations with OpenFOAM
Let’s suppose that we performed some Computational Fluid Dynamics (CFD) simulations using the open-source library OpenFOAM. This involves setting up a case with some initial conditions, running the simulations, post-processing the simulation data with a program like ParaView, and then post-processing that data with python.
Most research can be broken down into three processing stages: Pre-process, pProcess, Ppost-process. We will map the case study to these three as follows:
- Pre-process: generating the geometry, setting up the case files, determining which model to use, etc.
- Process: performing the simulation, generating the field data for each cell in the mesh, making sure the simulation converges, etc.
- Post-process: Convert all the field variables for each cell into a .csv file, use python to process .csv files to generate data and figures for publication, etc.
Let’s say we’ve run our simulations, generated our data and figures for publication, and now we need to make everything publicly available. There is a problem here though since our data is 1 TB, but our repository can only be 100 GB. Even after compressing our data to something like a .zip file, it is still 600 GB. On top of this requirement, there are physical limitations of the speed at which data can be downloaded so care must be taken to keep repositories as small as possible. So what data is important enough to keep in the repository. The data breakdown is as follows:
- Pre-processing: ~1 GB
- Processing: ~800 GB
- Post-processing: ~200 GB
We want to provide the readers with the raw data and the code that we used to post-process that raw data. This means we include all the data and code generated in the post-processing step. We can compress this down to ~50 GB. What about the pre-processing and processing steps?
Here we notice that the bulk of our data is in the processing step (~800 GB). But we note that by running the processing step with the data in the pre-processing step will generate the same data. In cases like this, we don’t need to include the data for the processing step, only the pre-processing data. A reader can regenerate our process data if they want. After compressing all this data, we get a repository from 1 TB down to ~50 GB.
A good data repository has enough data to reproduce the results. Your research data should be kept at an appropriate level of ‘rawness’ that allows it to be reused, and as small as possible to facilitate easy storage and data egress.
Only data that is needed to recreate the study and results should be included, and care must be taken to exclude data that may not be immediately needed to reproduce the results.
Key Points
- An important part of doing research is sharing it with others so that they can use it.
- Choose a repository that is field specific if you can, but generalist ones work too.
- Make sure to think about what data you actual need to share with other researchers so that they can recreate your results (does it need to be a 4 TB dataset?)
PLOS ONE. Recommended Repositories, 2023. URL https://journals.plos.org/ plosone/s/recommended-repositories.↩︎
NIH. Generalist Repositories, 2023. URL https://sharing.nih.gov/datamanagement- and-sharing-policy/sharing-scientific-data/generalistrepositories.↩︎
NIH. Generalist Repositories, 2023. URL https://sharing.nih.gov/datamanagement- and-sharing-policy/sharing-scientific-data/generalistrepositories.↩︎
Mark D. Wilkinson, Michel Dumontier, IJsbrand Jan Aalbersberg, Gabrielle Appleton, Myles Axton, Arie Baak, Niklas Blomberg, Jan-Willem Boiten, Luiz Bonino da Silva Santos, Philip E. Bourne, Jildau Bouwman, Anthony J. Brookes, Tim Clark, Mercè Crosas, Ingrid Dillo, Olivier Dumon, Scott Edmunds, Chris T. Evelo, Richard Finkers, Alejandra Gonzalez-Beltran, Alasdair J. G. Gray, Paul Groth, Carole Goble, Jeffrey S. Grethe, Jaap Heringa, Peter A. C. ’t Hoen, Rob Hooft, Tobias Kuhn, Ruben Kok, Joost Kok, Scott J. Lusher, Maryann E. Martone, Albert Mons, Abel L. Packer, Bengt Persson, Philippe Rocca-Serra, Marco Roos, Rene van Schaik, Susanna-Assunta Sansone, Erik Schultes, Thierry Sengstag, Ted Slater, George Strawn, Morris A. Swertz, Mark Thompson, Johan van der Lei, Erik van Mulligen, Jan Velterop, Andra Waagmeester, Peter Wittenburg, Katherine Wolstencroft, Jun Zhao, and Barend Mons. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data, 3(1):160018, March 2016. ISSN 2052-4463. doi: 10.1038/sdata.2016.18. URL https: //www.nature.com/articles/sdata201618. Number: 1 Publisher: Nature Publishing Group.↩︎
Creative Commons Wiki. CC0 use for data, 2014. URL https://wiki. creativecommons.org/wiki/CC0_use_for_data.↩︎
Scientific Data Data Policies. Nature 2024. URL https://www.nature.com/sdata/policies/ data-policies↩︎
